Differential gene expression analysis of HNSCC tumors deciphered tobacco dependent and independent molecular signatures
- PMID: 31692905
- PMCID: PMC6817442
- DOI: 10.18632/oncotarget.27249
Differential gene expression analysis of HNSCC tumors deciphered tobacco dependent and independent molecular signatures
Abstract
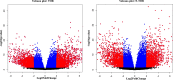
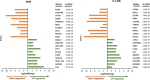
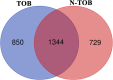
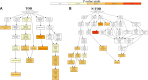
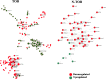
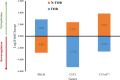
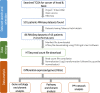
Head and neck cancer is the sixth most common cancer worldwide, with tobacco as the leading cause. However, it is increasing in non-tobacco users also, hence limiting our understanding of its underlying molecular mechanisms. RNA-seq analysis of cancers has proven as effective tool in understanding disease etiology. In the present study, RNA-Seq of 86 matched Tumor/Normal pairs, of tobacco smoking (TOB) and non-smokers (N-TOB) HNSCC samples analyzed, followed by validation on 375 similar datasets. Total 2194 and 2073 differentially expressed genes were identified in TOB and N-TOB tumors, respectively. GO analysis found muscle contraction as the most enriched biological process in both TOB and N-TOB tumors. Pathway analysis identified muscle contraction and salivary secretion pathways enriched in both categories, whereas calcium signaling and neuroactive ligand-receptor pathway was more enriched in TOB and N-TOB tumors respectively. Network analysis identified muscle development related genes as hub node i. e. ACTN2, MYL2 and TTN in both TOB and N-TOB tumors, whereas EGFR and MYH6, depicts specific role in TOB and N-TOB tumors. Additionally, we found enriched gene networks possibly be regulated by tumor suppressor miRNAs such as hsa-miR-29/a/b/c, hsa-miR-26b-5p etc., suggestive to be key riboswitches in regulatory cascade of HNSCC. Interestingly, three genes PKLR, CST1 and C17orf77 found to show opposite regulation in each category, hence suggested to be key genes in separating TOB from N-TOB tumors. Our investigation identified key genes involved in important pathways implicated in tobacco dependent and independent carcinogenesis hence may help in designing precise HNSCC diagnostics and therapeutics strategies.
Keywords: differentially expressed genes; head and neck squamous cell carcinoma (HNSCC); hub gene; miRNA; tobacco.
Copyright: © 2019 Shaikh et al.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no competing interests.
Figures

Similar articles
-
The Expression and Effection of MicroRNA-499a in High-Tobacco Exposed Head and Neck Squamous Cell Carcinoma: A Bioinformatic Analysis.Front Oncol. 2019 Jul 31;9:678. doi: 10.3389/fonc.2019.00678. eCollection 2019. Front Oncol. 2019. PMID: 31417866 Free PMC article.
-
Integrated analysis of deregulation microRNA expression in head and neck squamous cell carcinoma.Medicine (Baltimore). 2021 Feb 12;100(6):e24618. doi: 10.1097/MD.0000000000024618. Medicine (Baltimore). 2021. PMID: 33578572 Free PMC article.
-
Construction of the miRNA-mRNA regulatory network and analysis of hub genes in oral squamous cell carcinoma.Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2022 Sep;166(3):280-289. doi: 10.5507/bp.2022.001. Epub 2022 Feb 3. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2022. PMID: 35132271
-
Identification and Confirmation of the miR-30 Family as a Potential Central Player in Tobacco-Related Head and Neck Squamous Cell Carcinoma.Front Oncol. 2021 Jul 13;11:616372. doi: 10.3389/fonc.2021.616372. eCollection 2021. Front Oncol. 2021. PMID: 34336638 Free PMC article.
-
Tobacco-related carcinogenesis in head and neck cancer.Cancer Metastasis Rev. 2017 Sep;36(3):411-423. doi: 10.1007/s10555-017-9689-6. Cancer Metastasis Rev. 2017. PMID: 28801840 Free PMC article. Review.
Cited by
-
Novel Therapies for Tongue Squamous Cell Carcinoma Patients with High-Grade Tumors.Life (Basel). 2021 Aug 10;11(8):813. doi: 10.3390/life11080813. Life (Basel). 2021. PMID: 34440557 Free PMC article.
-
Identification of microRNAs Targeting the Transporter Associated with Antigen Processing TAP1 in Melanoma.J Clin Med. 2020 Aug 20;9(9):2690. doi: 10.3390/jcm9092690. J Clin Med. 2020. PMID: 32825219 Free PMC article.
-
MYL5 as a Novel Prognostic Marker is Associated with Immune Infiltrating in Breast Cancer: A Preliminary Study.Breast J. 2023 Feb 17;2023:9508632. doi: 10.1155/2023/9508632. eCollection 2023. Breast J. 2023. PMID: 36846347 Free PMC article.
-
A pan-cancer analysis of pituitary tumor-transforming 3, pseudogene.Am J Transl Res. 2023 Aug 15;15(8):5408-5424. eCollection 2023. Am J Transl Res. 2023. PMID: 37692950 Free PMC article.
-
Cystatin-B Negatively Regulates the Malignant Characteristics of Oral Squamous Cell Carcinoma Possibly Via the Epithelium Proliferation/Differentiation Program.Front Oncol. 2021 Aug 24;11:707066. doi: 10.3389/fonc.2021.707066. eCollection 2021. Front Oncol. 2021. PMID: 34504787 Free PMC article.
References
-
- Fukumoto I, Hanazawa T, Kinoshita T, Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer. 2015; 112:891–900. 10.1038/bjc.2015.19. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

