Split intein-mediated selection of cells containing two plasmids using a single antibiotic
- PMID: 31672972
- PMCID: PMC6823396
- DOI: 10.1038/s41467-019-12911-1
Split intein-mediated selection of cells containing two plasmids using a single antibiotic
Erratum in
-
Author Correction: Split intein-mediated selection of cells containing two plasmids using a single antibiotic.Nat Commun. 2020 Jan 9;11(1):276. doi: 10.1038/s41467-019-13716-y. Nat Commun. 2020. PMID: 31932594 Free PMC article.
Abstract
To build or dissect complex pathways in bacteria and mammalian cells, it is often necessary to recur to at least two plasmids, for instance harboring orthogonal inducible promoters. Here we present SiMPl, a method based on rationally designed split enzymes and intein-mediated protein trans-splicing, allowing the selection of cells carrying two plasmids with a single antibiotic. We show that, compared to the traditional method based on two antibiotics, SiMPl increases the production of the antimicrobial non-ribosomal peptide indigoidine and the non-proteinogenic aromatic amino acid para-amino-L-phenylalanine from bacteria. Using a human T cell line, we employ SiMPl to obtain a highly pure population of cells double positive for the two chains of the T cell receptor, TCRα and TCRβ, using a single antibiotic. SiMPl has profound implications for metabolic engineering and for constructing complex synthetic circuits in bacteria and mammalian cells.
Conflict of interest statement
The authors declare no competing interests.
Figures
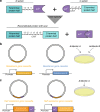
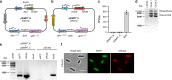

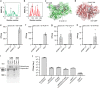

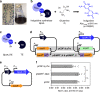



Similar articles
-
Split selectable markers.Nat Commun. 2019 Oct 31;10(1):4968. doi: 10.1038/s41467-019-12891-2. Nat Commun. 2019. PMID: 31672965 Free PMC article.
-
Protein trans-splicing on an M13 bacteriophage: towards directed evolution of a semisynthetic split intein by phage display.J Pept Sci. 2010 Oct;16(10):575-81. doi: 10.1002/psc.1243. J Pept Sci. 2010. PMID: 20862725
-
Trans protein splicing of cyanobacterial split inteins in endogenous and exogenous combinations.Biochemistry. 2007 Jan 9;46(1):322-30. doi: 10.1021/bi0611762. Biochemistry. 2007. PMID: 17198403
-
Protein trans-splicing and its use in structural biology: opportunities and limitations.Mol Biosyst. 2010 Nov;6(11):2110-21. doi: 10.1039/c0mb00034e. Epub 2010 Aug 31. Mol Biosyst. 2010. PMID: 20820635 Review.
-
Split-inteins and their bioapplications.Biotechnol Lett. 2015 Nov;37(11):2121-37. doi: 10.1007/s10529-015-1905-2. Epub 2015 Jul 8. Biotechnol Lett. 2015. PMID: 26153348 Review.
Cited by
-
Thermally controlled intein splicing of engineered DNA polymerases provides a robust and generalizable solution for accurate and sensitive molecular diagnostics.Nucleic Acids Res. 2023 Jun 23;51(11):5883-5894. doi: 10.1093/nar/gkad368. Nucleic Acids Res. 2023. PMID: 37166959 Free PMC article.
-
Int&in: A machine learning-based web server for active split site identification in inteins.Protein Sci. 2024 Jun;33(6):e4985. doi: 10.1002/pro.4985. Protein Sci. 2024. PMID: 38717278 Free PMC article.
-
A designed fusion tag for soluble expression and selective separation of extracellular domains of fibroblast growth factor receptors.Sci Rep. 2021 Nov 2;11(1):21453. doi: 10.1038/s41598-021-01029-4. Sci Rep. 2021. PMID: 34728710 Free PMC article.
-
The expanding role of split protein complementation in opsin-free optogenetics.Curr Opin Pharmacol. 2022 Aug;65:102236. doi: 10.1016/j.coph.2022.102236. Epub 2022 May 21. Curr Opin Pharmacol. 2022. PMID: 35609383 Free PMC article. Review.
-
Expanding the SiMPl Plasmid Toolbox for Use with Spectinomycin/Streptomycin.ACS Omega. 2021 May 21;6(22):14148-14153. doi: 10.1021/acsomega.1c00649. eCollection 2021 Jun 8. ACS Omega. 2021. PMID: 34124437 Free PMC article.
References
-
- Hirata R, et al. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:6726–6733. - PubMed
-
- Kane PM, et al. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science. 1990;250:651–657. - PubMed
-
- Brenzel S, Kurpiers T, Mootz HD. Engineering artificially split inteins for applications in protein chemistry: biochemical characterization of the split Ssp DnaB intein and comparison to the split Sce VMA intein. Biochemistry. 2006;45:1571–1578. - PubMed
-
- Zettler J, Schutz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. - PubMed
-
- Siu KH, Chen W. Control of the Yeast mating pathway by reconstitution of functional alpha-factor using split intein-catalyzed reactions. ACS Synth. Biol. 2017;6:1453–1460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

