Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA
- PMID: 31671674
- PMCID: PMC6895833
- DOI: 10.3390/genes10110868
Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA
Abstract
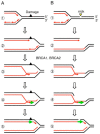
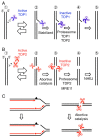
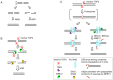
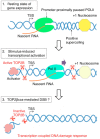
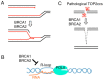
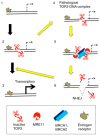
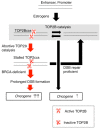
Type II DNA topoisomerase enzymes (TOP2) catalyze topological changes by strand passage reactions. They involve passing one intact double stranded DNA duplex through a transient enzyme-bridged break in another (gated helix) followed by ligation of the break by TOP2. A TOP2 poison, etoposide blocks TOP2 catalysis at the ligation step of the enzyme-bridged break, increasing the number of stable TOP2 cleavage complexes (TOP2ccs). Remarkably, such pathological TOP2ccs are formed during the normal cell cycle as well as in postmitotic cells. Thus, this 'abortive catalysis' can be a major source of spontaneously arising DNA double-strand breaks (DSBs). TOP2-mediated DSBs are also formed upon stimulation with physiological concentrations of androgens and estrogens. The frequent occurrence of TOP2-mediated DSBs was previously not appreciated because they are efficiently repaired. This repair is performed in collaboration with BRCA1, BRCA2, MRE11 nuclease, and tyrosyl-DNA phosphodiesterase 2 (TDP2) with nonhomologous end joining (NHEJ) factors. This review first discusses spontaneously arising DSBs caused by the abortive catalysis of TOP2 and then summarizes proteins involved in repairing stalled TOP2ccs and discusses the genotoxicity of the sex hormones.
Keywords: BRCA1; BRCA2; Topoisomerase II; breast cancer; cell cycle; estrogen; genotoxicity; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
TDP2 suppresses genomic instability induced by androgens in the epithelial cells of prostate glands.Genes Cells. 2020 Jul;25(7):450-465. doi: 10.1111/gtc.12770. Epub 2020 May 5. Genes Cells. 2020. PMID: 32277721 Free PMC article.
-
TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription.Nat Commun. 2017 Aug 10;8(1):233. doi: 10.1038/s41467-017-00307-y. Nat Commun. 2017. PMID: 28794467 Free PMC article.
-
TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo.PLoS Genet. 2013;9(3):e1003226. doi: 10.1371/journal.pgen.1003226. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505375 Free PMC article.
-
End-processing nucleases and phosphodiesterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair.DNA Repair (Amst). 2016 Jul;43:57-68. doi: 10.1016/j.dnarep.2016.05.011. Epub 2016 May 18. DNA Repair (Amst). 2016. PMID: 27262532 Review.
-
Mechanisms to Repair Stalled Topoisomerase II-DNA Covalent Complexes.Mol Pharmacol. 2022 Jan;101(1):24-32. doi: 10.1124/molpharm.121.000374. Epub 2021 Oct 23. Mol Pharmacol. 2022. PMID: 34689119 Review.
Cited by
-
Expression of the Topoisomerase II Alpha (TOP2A) Gene in Lung Adenocarcinoma Cells and the Association with Patient Outcomes.Med Sci Monit. 2020 Dec 27;26:e929120. doi: 10.12659/MSM.929120. Med Sci Monit. 2020. PMID: 33361736 Free PMC article.
-
Targeting DNA Damage Response in Prostate and Breast Cancer.Int J Mol Sci. 2020 Nov 4;21(21):8273. doi: 10.3390/ijms21218273. Int J Mol Sci. 2020. PMID: 33158305 Free PMC article. Review.
-
Evaluation of the Effect of Dibornol on the Level of DNA Damage in the DNA Comet Test In Vivo.Bull Exp Biol Med. 2024 Sep;177(5):635-638. doi: 10.1007/s10517-024-06239-0. Epub 2024 Sep 28. Bull Exp Biol Med. 2024. PMID: 39340622
-
Cross-species incompatibility between a DNA satellite and the Drosophila Spartan homolog poisons germline genome integrity.Curr Biol. 2022 Jul 11;32(13):2962-2971.e4. doi: 10.1016/j.cub.2022.05.009. Epub 2022 May 27. Curr Biol. 2022. PMID: 35643081 Free PMC article.
-
Checkpoint kinase‑1 inhibition and etoposide exhibit a strong synergistic anticancer effect on chronic myeloid leukemia cell line K562 by impairing homologous recombination DNA damage repair.Oncol Rep. 2020 Nov;44(5):2152-2164. doi: 10.3892/or.2020.7757. Epub 2020 Sep 7. Oncol Rep. 2020. PMID: 32901871 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

