ER-phagy and human diseases
- PMID: 31659280
- PMCID: PMC7206075
- DOI: 10.1038/s41418-019-0444-0
ER-phagy and human diseases
Abstract
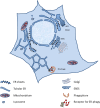
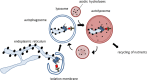
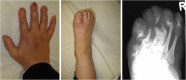
Autophagy regulates the degradation of unnecessary or dysfunctional cellular components. This catabolic process requires the formation of a double-membrane vesicle, the autophagosome, that engulfs the cytosolic material and delivers it to the lysosome. Substrate specificity is achieved by autophagy receptors, which are characterized by the presence of at least one LC3-interaction region (LIR) or GABARAP-interaction motif (GIM). Only recently, several receptors that mediate the specific degradation of endoplasmic reticulum (ER) components via autophagy have been identified (the process known as ER-phagy or reticulophagy). Here, we give an update on the current knowledge about the role of ER-phagy receptors in health and disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
ER-phagy: mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum.Physiol Rev. 2022 Jul 1;102(3):1393-1448. doi: 10.1152/physrev.00038.2021. Epub 2022 Feb 21. Physiol Rev. 2022. PMID: 35188422 Free PMC article. Review.
-
ER-Phagy, ER Homeostasis, and ER Quality Control: Implications for Disease.Trends Biochem Sci. 2021 Aug;46(8):630-639. doi: 10.1016/j.tibs.2020.12.013. Epub 2021 Jan 25. Trends Biochem Sci. 2021. PMID: 33509650 Free PMC article. Review.
-
Eat it right: ER-phagy and recovER-phagy.Biochem Soc Trans. 2018 Jun 19;46(3):699-706. doi: 10.1042/BST20170354. Epub 2018 May 25. Biochem Soc Trans. 2018. PMID: 29802216 Free PMC article. Review.
-
ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy.Curr Biol. 2019 Mar 4;29(5):846-855.e6. doi: 10.1016/j.cub.2019.01.041. Epub 2019 Feb 14. Curr Biol. 2019. PMID: 30773365
-
Advances in ER-Phagy and Its Diseases Relevance.Cells. 2021 Sep 6;10(9):2328. doi: 10.3390/cells10092328. Cells. 2021. PMID: 34571977 Free PMC article. Review.
Cited by
-
Quantitative and time-resolved monitoring of organelle and protein delivery to the lysosome with a tandem fluorescent Halo-GFP reporter.Mol Biol Cell. 2022 May 15;33(6):ar57. doi: 10.1091/mbc.E21-10-0526. Epub 2022 Feb 2. Mol Biol Cell. 2022. PMID: 35108065 Free PMC article.
-
MEF2D-NR4A1-FAM134B2-mediated reticulophagy contributes to amino acid homeostasis.Autophagy. 2022 May;18(5):1049-1061. doi: 10.1080/15548627.2021.1968228. Epub 2021 Sep 14. Autophagy. 2022. PMID: 34517786 Free PMC article.
-
Autophagy, a double-edged sword for oral tissue regeneration.J Adv Res. 2024 May;59:141-159. doi: 10.1016/j.jare.2023.06.010. Epub 2023 Jun 24. J Adv Res. 2024. PMID: 37356803 Free PMC article. Review.
-
Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18530-18539. doi: 10.1073/pnas.2008923117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690699 Free PMC article.
-
ER remodeling via ER-phagy.Mol Cell. 2022 Apr 21;82(8):1492-1500. doi: 10.1016/j.molcel.2022.02.018. Mol Cell. 2022. PMID: 35452617 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

