53BP1 Enforces Distinct Pre- and Post-resection Blocks on Homologous Recombination
- PMID: 31653568
- PMCID: PMC6993210
- DOI: 10.1016/j.molcel.2019.09.024
53BP1 Enforces Distinct Pre- and Post-resection Blocks on Homologous Recombination
Abstract
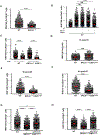
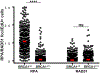
53BP1 activity drives genome instability and lethality in BRCA1-deficient mice by inhibiting homologous recombination (HR). The anti-recombinogenic functions of 53BP1 require phosphorylation-dependent interactions with PTIP and RIF1/shieldin effector complexes. While RIF1/shieldin blocks 5'-3' nucleolytic processing of DNA ends, it remains unclear how PTIP antagonizes HR. Here, we show that mutation of the PTIP interaction site in 53BP1 (S25A) allows sufficient DNA2-dependent end resection to rescue the lethality of BRCA1Δ11 mice, despite increasing RIF1 "end-blocking" at DNA damage sites. However, double-mutant cells fail to complete HR, as excessive shieldin activity also inhibits RNF168-mediated loading of PALB2/RAD51. As a result, BRCA1Δ1153BP1S25A mice exhibit hallmark features of HR insufficiency, including premature aging and hypersensitivity to PARPi. Disruption of shieldin or forced targeting of PALB2 to ssDNA in BRCA1D1153BP1S25A cells restores RNF168 recruitment, RAD51 nucleofilament formation, and PARPi resistance. Our study therefore reveals a critical function of shieldin post-resection that limits the loading of RAD51.
Keywords: 53BP1; BRCA1; PARPi; aging; cancer; homologous recombination; resection; shieldin.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures

Similar articles
-
BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation.Mol Cell. 2019 Mar 21;73(6):1267-1281.e7. doi: 10.1016/j.molcel.2018.12.010. Epub 2019 Jan 28. Mol Cell. 2019. PMID: 30704900 Free PMC article.
-
53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polα-primase fill-in synthesis.Nat Cell Biol. 2022 Jan;24(1):51-61. doi: 10.1038/s41556-021-00812-9. Epub 2022 Jan 13. Nat Cell Biol. 2022. PMID: 35027730 Free PMC article.
-
53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in.Nature. 2018 Aug;560(7716):112-116. doi: 10.1038/s41586-018-0324-7. Epub 2018 Jul 18. Nature. 2018. PMID: 30022158 Free PMC article.
-
Shieldin - the protector of DNA ends.EMBO Rep. 2019 May;20(5):e47560. doi: 10.15252/embr.201847560. Epub 2019 Apr 4. EMBO Rep. 2019. PMID: 30948458 Free PMC article. Review.
-
The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication.Nat Rev Mol Cell Biol. 2020 May;21(5):284-299. doi: 10.1038/s41580-020-0218-z. Epub 2020 Feb 24. Nat Rev Mol Cell Biol. 2020. PMID: 32094664 Free PMC article. Review.
Cited by
-
Centromeres under Pressure: Evolutionary Innovation in Conflict with Conserved Function.Genes (Basel). 2020 Aug 10;11(8):912. doi: 10.3390/genes11080912. Genes (Basel). 2020. PMID: 32784998 Free PMC article. Review.
-
Rad9/53BP1 promotes DNA repair via crossover recombination by limiting the Sgs1 and Mph1 helicases.Nat Commun. 2020 Jun 23;11(1):3181. doi: 10.1038/s41467-020-16997-w. Nat Commun. 2020. PMID: 32576832 Free PMC article.
-
Fighting resistance: post-PARP inhibitor treatment strategies in ovarian cancer.Ther Adv Med Oncol. 2023 Mar 1;15:17588359231157644. doi: 10.1177/17588359231157644. eCollection 2023. Ther Adv Med Oncol. 2023. PMID: 36872947 Free PMC article. Review.
-
RNF168-mediated localization of BARD1 recruits the BRCA1-PALB2 complex to DNA damage.Nat Commun. 2021 Aug 18;12(1):5016. doi: 10.1038/s41467-021-25346-4. Nat Commun. 2021. PMID: 34408138 Free PMC article.
-
The chromatin-associated 53BP1 ortholog, HSR-9, regulates recombinational repair and X chromosome segregation in the Caenorhabditis elegans germ line.bioRxiv [Preprint]. 2024 Apr 15:2024.04.12.589267. doi: 10.1101/2024.04.12.589267. bioRxiv. 2024. Update in: Genetics. 2024 Aug 7;227(4):iyae102. doi: 10.1093/genetics/iyae102. PMID: 38659880 Free PMC article. Updated. Preprint.
References
-
- Baer R, and Ludwig T (2002). The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev 12, 86–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

