Search-and-replace genome editing without double-strand breaks or donor DNA
- PMID: 31634902
- PMCID: PMC6907074
- DOI: 10.1038/s41586-019-1711-4
Search-and-replace genome editing without double-strand breaks or donor DNA
Abstract
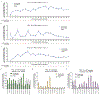
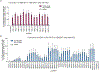
Most genetic variants that contribute to disease1 are challenging to correct efficiently and without excess byproducts2-5. Here we describe prime editing, a versatile and precise genome editing method that directly writes new genetic information into a specified DNA site using a catalytically impaired Cas9 endonuclease fused to an engineered reverse transcriptase, programmed with a prime editing guide RNA (pegRNA) that both specifies the target site and encodes the desired edit. We performed more than 175 edits in human cells, including targeted insertions, deletions, and all 12 types of point mutation, without requiring double-strand breaks or donor DNA templates. We used prime editing in human cells to correct, efficiently and with few byproducts, the primary genetic causes of sickle cell disease (requiring a transversion in HBB) and Tay-Sachs disease (requiring a deletion in HEXA); to install a protective transversion in PRNP; and to insert various tags and epitopes precisely into target loci. Four human cell lines and primary post-mitotic mouse cortical neurons support prime editing with varying efficiencies. Prime editing shows higher or similar efficiency and fewer byproducts than homology-directed repair, has complementary strengths and weaknesses compared to base editing, and induces much lower off-target editing than Cas9 nuclease at known Cas9 off-target sites. Prime editing substantially expands the scope and capabilities of genome editing, and in principle could correct up to 89% of known genetic variants associated with human diseases.
Figures










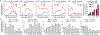
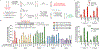

Comment in
-
Super-precise new CRISPR tool could tackle a plethora of genetic diseases.Nature. 2019 Oct;574(7779):464-465. doi: 10.1038/d41586-019-03164-5. Nature. 2019. PMID: 31641267 No abstract available.
-
A prime alternative.Nat Rev Genet. 2020 Jan;21(1):3. doi: 10.1038/s41576-019-0191-6. Nat Rev Genet. 2020. PMID: 31649330 No abstract available.
-
CRISPR-Cas in its prime.Nat Rev Mol Cell Biol. 2019 Dec;20(12):718-719. doi: 10.1038/s41580-019-0194-3. Nat Rev Mol Cell Biol. 2019. PMID: 31685954 No abstract available.
-
Human germline editing needs one message.Nature. 2019 Nov;575(7783):415-416. doi: 10.1038/d41586-019-03525-0. Nature. 2019. PMID: 31748735 No abstract available.
-
Expanding the gene editing landscape.Nat Rev Drug Discov. 2019 Nov;18(12):904. doi: 10.1038/d41573-019-00178-6. Nat Rev Drug Discov. 2019. PMID: 31780843 No abstract available.
-
CRISPR tool modifies genes precisely by copying RNA into the genome.Nature. 2019 Dec;576(7785):48-49. doi: 10.1038/d41586-019-03392-9. Nature. 2019. PMID: 31792413 No abstract available.
-
Uncut but Primed for Change.CRISPR J. 2019 Dec;2(6):352-354. doi: 10.1089/crispr.2019.29079.mji. CRISPR J. 2019. PMID: 31860347 No abstract available.
-
Quest to use CRISPR against disease gains ground.Nature. 2020 Jan;577(7789):156. doi: 10.1038/d41586-019-03919-0. Nature. 2020. PMID: 31911695 No abstract available.
-
Prime Editing: Precision Genome Editing by Reverse Transcription.Mol Cell. 2020 Jan 16;77(2):210-212. doi: 10.1016/j.molcel.2019.12.016. Mol Cell. 2020. PMID: 31951546
-
Prime Editing: Game Changer for Modifying Plant Genomes.Trends Plant Sci. 2020 Aug;25(8):722-724. doi: 10.1016/j.tplants.2020.05.008. Epub 2020 Jun 5. Trends Plant Sci. 2020. PMID: 32513585
Similar articles
-
Highly efficient generation of isogenic pluripotent stem cell models using prime editing.Elife. 2022 Sep 7;11:e79208. doi: 10.7554/eLife.79208. Elife. 2022. PMID: 36069759 Free PMC article.
-
Prime Editing: A New Way for Genome Editing.Trends Cell Biol. 2020 Apr;30(4):257-259. doi: 10.1016/j.tcb.2020.01.004. Epub 2020 Jan 27. Trends Cell Biol. 2020. PMID: 32001098
-
Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage.Nature. 2016 May 19;533(7603):420-4. doi: 10.1038/nature17946. Epub 2016 Apr 20. Nature. 2016. PMID: 27096365 Free PMC article.
-
Prime editing for precise and highly versatile genome manipulation.Nat Rev Genet. 2023 Mar;24(3):161-177. doi: 10.1038/s41576-022-00541-1. Epub 2022 Nov 7. Nat Rev Genet. 2023. PMID: 36344749 Free PMC article. Review.
-
[Prime-Editing Methods and pegRNA Design Programs].Mol Biol (Mosk). 2024 Jan-Feb;58(1):22-39. Mol Biol (Mosk). 2024. PMID: 38943578 Review. Russian.
Cited by
-
Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi.Front Microbiol. 2021 Feb 11;12:638096. doi: 10.3389/fmicb.2021.638096. eCollection 2021. Front Microbiol. 2021. PMID: 33643273 Free PMC article. Review.
-
Sequence modification on demand: search and replace tools for precise gene editing in plants.Transgenic Res. 2021 Aug;30(4):353-379. doi: 10.1007/s11248-021-00253-y. Epub 2021 Jun 4. Transgenic Res. 2021. PMID: 34086167 Review.
-
Translational potential of base-editing tools for gene therapy of monogenic diseases.Front Bioeng Biotechnol. 2022 Aug 10;10:942440. doi: 10.3389/fbioe.2022.942440. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36032737 Free PMC article. Review.
-
Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins.Nat Commun. 2021 Mar 2;12(1):1384. doi: 10.1038/s41467-021-21559-9. Nat Commun. 2021. PMID: 33654077 Free PMC article.
-
Optimized prime editing in monocot plants using PlantPegDesigner and engineered plant prime editors (ePPEs).Nat Protoc. 2023 Mar;18(3):831-853. doi: 10.1038/s41596-022-00773-9. Epub 2022 Nov 25. Nat Protoc. 2023. PMID: 36434096 Review.
References
Methods References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

