Remove, Recycle, Degrade: Regulating Plasma Membrane Protein Accumulation
- PMID: 31628169
- PMCID: PMC6925004
- DOI: 10.1105/tpc.19.00433
Remove, Recycle, Degrade: Regulating Plasma Membrane Protein Accumulation
Abstract
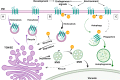
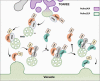
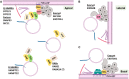
Interactions between plant cells and the environment rely on modulation of protein receptors, transporters, channels, and lipids at the plasma membrane (PM) to facilitate intercellular communication, nutrient uptake, environmental sensing, and directional growth. These functions are fine-tuned by cellular pathways maintaining or reducing particular proteins at the PM. Proteins are endocytosed, and their fate is decided between recycling and degradation to modulate localization, abundance, and activity. Selective autophagy is another pathway regulating PM protein accumulation in response to specific conditions or developmental signals. The mechanisms regulating recycling, degradation, and autophagy have been studied extensively, yet we are just now addressing their regulation and coordination. Here, we (1) provide context concerning regulation of protein accumulation, recycling, or degradation by overviewing endomembrane trafficking; (2) discuss pathways regulating recycling and degradation in terms of cellular roles and cargoes; (3) review plant selective autophagy and its physiological significance; (4) focus on two decision-making mechanisms: regulation of recycling versus degradation of PM proteins and coordination between autophagy and vacuolar degradation; and (5) identify future challenges.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures



Similar articles
-
A plant-unique protein BLISTER coordinates with core retromer to modulate endosomal sorting of plasma membrane and vacuolar proteins.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2211258120. doi: 10.1073/pnas.2211258120. Epub 2022 Dec 28. Proc Natl Acad Sci U S A. 2023. PMID: 36577063 Free PMC article.
-
Endosomal functions in plants.Traffic. 2008 Sep;9(10):1589-98. doi: 10.1111/j.1600-0854.2008.00787.x. Epub 2008 Jul 9. Traffic. 2008. PMID: 18627577 Review.
-
Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway.BMC Plant Biol. 2012 Sep 12;12:164. doi: 10.1186/1471-2229-12-164. BMC Plant Biol. 2012. PMID: 22970698 Free PMC article.
-
Knowns and unknowns of plasma membrane protein degradation in plants.Plant Sci. 2018 Jul;272:55-61. doi: 10.1016/j.plantsci.2018.04.008. Epub 2018 Apr 12. Plant Sci. 2018. PMID: 29807606 Review.
-
Dynamic reorganization of the endomembrane system during spermatogenesis in Marchantia polymorpha.J Plant Res. 2017 May;130(3):433-441. doi: 10.1007/s10265-017-0909-5. Epub 2017 Feb 3. J Plant Res. 2017. PMID: 28160149
Cited by
-
Fluid-phase and membrane markers reveal spatio-temporal dynamics of membrane traffic and repair in the green alga Chara australis.Protoplasma. 2021 Jul;258(4):711-728. doi: 10.1007/s00709-021-01627-z. Epub 2021 Mar 11. Protoplasma. 2021. PMID: 33704568 Free PMC article.
-
Subcellular trafficking and post-translational modification regulate PIN polarity in plants.Front Plant Sci. 2022 Jul 27;13:923293. doi: 10.3389/fpls.2022.923293. eCollection 2022. Front Plant Sci. 2022. PMID: 35968084 Free PMC article. Review.
-
The plant-unique protein DRIF1 coordinates with sorting nexin 1 to regulate membrane protein homeostasis.Plant Cell. 2023 Nov 30;35(12):4217-4237. doi: 10.1093/plcell/koad227. Plant Cell. 2023. PMID: 37647529 Free PMC article.
-
The Arabidopsis SAC9 enzyme is enriched in a cortical population of early endosomes and restricts PI(4,5)P2 at the plasma membrane.Elife. 2022 Aug 31;11:e73837. doi: 10.7554/eLife.73837. Elife. 2022. PMID: 36044021 Free PMC article.
-
Rice STOMATAL CYTOKINESIS DEFECTIVE2 regulates cell expansion by affecting vesicular trafficking in rice.Plant Physiol. 2022 Jun 1;189(2):567-584. doi: 10.1093/plphys/kiac073. Plant Physiol. 2022. PMID: 35234957 Free PMC article.
References
-
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L.M., Catching A., Kirik V., Marc J., Overall R., Wasteneys G.O. (2013). CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 24: 649–659. - PubMed
-
- Assaad F.F., Qiu J.-L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129. - PMC - PubMed
-
- Avin-Wittenberg T., et al. (2018). Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69: 1335–1353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

