The Methyl-CpG-Binding Domain 2 and 3 Proteins and Formation of the Nucleosome Remodeling and Deacetylase Complex
- PMID: 31626804
- PMCID: PMC7156326
- DOI: 10.1016/j.jmb.2019.10.007
The Methyl-CpG-Binding Domain 2 and 3 Proteins and Formation of the Nucleosome Remodeling and Deacetylase Complex
Abstract
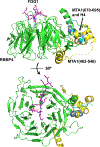
The Nucleosome Remodeling and Deacetylase (NuRD) complex uniquely combines both deacetylase and remodeling enzymatic activities in a single macromolecular complex. The methyl-CpG-binding domain 2 and 3 (MBD2 and MBD3) proteins provide a critical structural link between the deacetylase and remodeling components, while MBD2 endows the complex with the ability to selectively recognize methylated DNA. Hence, NuRD combines three major arms of epigenetic gene regulation. Research over the past few decades has revealed much of the structural basis driving formation of this complex and started to uncover the functional roles of NuRD in epigenetic gene regulation. However, we have yet to fully understand the molecular and biophysical basis for methylation-dependent chromatin remodeling and transcription regulation by NuRD. In this review, we discuss the structural information currently available for the complex, the role MBD2 and MBD3 play in forming and recruiting the complex to methylated DNA, and the biological functions of NuRD.
Keywords: Chromatin; DNA methylation; Epigenetics; Histone deacetylase; Nucleosome remodeling.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures
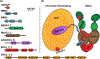

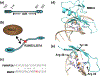

Similar articles
-
MBD3L2 interacts with MBD3 and components of the NuRD complex and can oppose MBD2-MeCP1-mediated methylation silencing.J Biol Chem. 2005 Apr 1;280(13):12700-9. doi: 10.1074/jbc.M413492200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15701600
-
Densely methylated DNA traps Methyl-CpG-binding domain protein 2 but permits free diffusion by Methyl-CpG-binding domain protein 3.J Biol Chem. 2022 Oct;298(10):102428. doi: 10.1016/j.jbc.2022.102428. Epub 2022 Aug 28. J Biol Chem. 2022. PMID: 36037972 Free PMC article.
-
Analysis of the complex between MBD2 and the histone deacetylase core of NuRD reveals key interactions critical for gene silencing.Proc Natl Acad Sci U S A. 2023 Aug 15;120(33):e2307287120. doi: 10.1073/pnas.2307287120. Epub 2023 Aug 8. Proc Natl Acad Sci U S A. 2023. PMID: 37552759 Free PMC article.
-
Emerging Molecular and Biological Functions of MBD2, a Reader of DNA Methylation.Front Genet. 2016 May 26;7:93. doi: 10.3389/fgene.2016.00093. eCollection 2016. Front Genet. 2016. PMID: 27303433 Free PMC article. Review.
-
The NuRD complex: linking histone modification to nucleosome remodeling.Curr Top Microbiol Immunol. 2003;274:269-90. doi: 10.1007/978-3-642-55747-7_10. Curr Top Microbiol Immunol. 2003. PMID: 12596911 Review.
Cited by
-
Identification of Novel MeCP2 Cancer-Associated Target Genes and Post-Translational Modifications.Front Oncol. 2020 Dec 10;10:576362. doi: 10.3389/fonc.2020.576362. eCollection 2020. Front Oncol. 2020. PMID: 33363010 Free PMC article.
-
BMP suppresses Wnt signaling via the Bcl11b-regulated NuRD complex to maintain intestinal stem cells.EMBO J. 2024 Dec;43(23):6032-6051. doi: 10.1038/s44318-024-00276-1. Epub 2024 Oct 21. EMBO J. 2024. PMID: 39433900 Free PMC article.
-
Increased Methyl-CpG-Binding Domain Protein 2 Promotes Cigarette Smoke-Induced Pulmonary Hypertension.Front Oncol. 2022 Jun 16;12:879793. doi: 10.3389/fonc.2022.879793. eCollection 2022. Front Oncol. 2022. PMID: 35785161 Free PMC article.
-
Making it or breaking it: DNA methylation and genome integrity.Essays Biochem. 2020 Oct 26;64(5):687-703. doi: 10.1042/EBC20200009. Essays Biochem. 2020. PMID: 32808652 Free PMC article. Review.
-
The topology of chromatin-binding domains in the NuRD deacetylase complex.Nucleic Acids Res. 2020 Dec 16;48(22):12972-12982. doi: 10.1093/nar/gkaa1121. Nucleic Acids Res. 2020. PMID: 33264408 Free PMC article.
References
-
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–77. - PubMed
-
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. - PubMed
-
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–6. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

