Chiral checkpoints during protein biosynthesis
- PMID: 31591268
- PMCID: PMC6851308
- DOI: 10.1074/jbc.REV119.008166
Chiral checkpoints during protein biosynthesis
Abstract
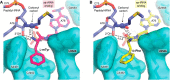
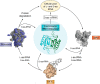
Protein chains contain only l-amino acids, with the exception of the achiral glycine, making the chains homochiral. This homochirality is a prerequisite for proper protein folding and, hence, normal cellular function. The importance of d-amino acids as a component of the bacterial cell wall and their roles in neurotransmission in higher eukaryotes are well-established. However, the wider presence and the corresponding physiological roles of these specific amino acid stereoisomers have been appreciated only recently. Therefore, it is expected that enantiomeric fidelity has to be a key component of all of the steps in translation. Cells employ various molecular mechanisms for keeping d-amino acids away from the synthesis of nascent polypeptide chains. The major factors involved in this exclusion are aminoacyl-tRNA synthetases (aaRSs), elongation factor thermo-unstable (EF-Tu), the ribosome, and d-aminoacyl-tRNA deacylase (DTD). aaRS, EF-Tu, and the ribosome act as "chiral checkpoints" by preferentially binding to l-amino acids or l-aminoacyl-tRNAs, thereby excluding d-amino acids. Interestingly, DTD, which is conserved across all life forms, performs "chiral proofreading," as it removes d-amino acids erroneously added to tRNA. Here, we comprehensively review d-amino acids with respect to their occurrence and physiological roles, implications for chiral checkpoints required for translation fidelity, and potential use in synthetic biology.
Keywords: D-amino acids; amino acid; aminoacyl tRNA synthetase; checkpoint control; chirality; genetic code; proofreading; proteins; ribosome; stereoselectivity; transfer RNA (tRNA); translation; translation elongation factor.
© 2019 Kuncha et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
Elongation Factor Tu Prevents Misediting of Gly-tRNA(Gly) Caused by the Design Behind the Chiral Proofreading Site of D-Aminoacyl-tRNA Deacylase.PLoS Biol. 2016 May 25;14(5):e1002465. doi: 10.1371/journal.pbio.1002465. eCollection 2016 May. PLoS Biol. 2016. PMID: 27224426 Free PMC article.
-
Chiral proofreading during protein biosynthesis and its evolutionary implications.FEBS Lett. 2022 Jul;596(13):1615-1627. doi: 10.1002/1873-3468.14419. Epub 2022 Jun 29. FEBS Lett. 2022. PMID: 35662005 Review.
-
Two proofreading steps amplify the accuracy of genetic code translation.Proc Natl Acad Sci U S A. 2016 Nov 29;113(48):13744-13749. doi: 10.1073/pnas.1610917113. Epub 2016 Nov 11. Proc Natl Acad Sci U S A. 2016. PMID: 27837019 Free PMC article.
-
Amino acid specificity in translation.Trends Biochem Sci. 2005 Dec;30(12):659-65. doi: 10.1016/j.tibs.2005.10.006. Epub 2005 Nov 2. Trends Biochem Sci. 2005. PMID: 16260144
-
Engineering Translation Components Improve Incorporation of Exotic Amino Acids.Int J Mol Sci. 2019 Jan 26;20(3):522. doi: 10.3390/ijms20030522. Int J Mol Sci. 2019. PMID: 30691159 Free PMC article. Review.
Cited by
-
The tRNA identity landscape for aminoacylation and beyond.Nucleic Acids Res. 2023 Feb 28;51(4):1528-1570. doi: 10.1093/nar/gkad007. Nucleic Acids Res. 2023. PMID: 36744444 Free PMC article. Review.
-
Study on serum metabolomics characteristics of obese patients with erectile dysfunction.Medicine (Baltimore). 2024 Oct 25;103(43):e40093. doi: 10.1097/MD.0000000000040093. Medicine (Baltimore). 2024. PMID: 39470567 Free PMC article.
-
A translation proofreader of archaeal origin imparts multi-aldehyde stress tolerance to land plants.Elife. 2024 Feb 19;12:RP92827. doi: 10.7554/eLife.92827. Elife. 2024. PMID: 38372335 Free PMC article.
-
Antibacterial Activity and Mechanism of Linalool against Shewanella putrefaciens.Molecules. 2021 Jan 5;26(1):245. doi: 10.3390/molecules26010245. Molecules. 2021. PMID: 33466475 Free PMC article.
-
DTD1 modulates synaptic efficacy by maintaining D-serine and D-aspartate homeostasis.Sci China Life Sci. 2025 Feb;68(2):467-483. doi: 10.1007/s11427-023-2681-y. Epub 2024 Oct 16. Sci China Life Sci. 2025. PMID: 39428430
References
-
- Pasteur L. (1848) Recherches sur les Relations qui peuvent Exister entre la Forme Cristalline, la Composition Chimique et le Sens de la Polarisation Rotatoire. Annales de Chimie et de Physique 24, 422–459
-
- Mahalakshmi R., and Balaram P. (2007) d-Amino Acids: A New Frontier in Amino Acid and Protein Research, pp. 415–430, Nova Science Publishers, Inc., Hauppauge, NY
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

