Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4+ T Cells
- PMID: 31578289
- PMCID: PMC6880164
- DOI: 10.1128/JVI.00969-19
Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4+ T Cells
Abstract
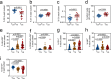
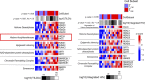
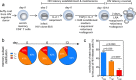
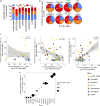
During antiretroviral therapy (ART), human immunodeficiency virus type 1 (HIV-1) persists as a latent reservoir in CD4+ T cell subsets in central memory (TCM), transitional memory (TTM), and effector memory (TEM) CD4+ T cells. We have identified differences in mechanisms underlying latency and responses to latency-reversing agents (LRAs) in ex vivo CD4+ memory T cells from virally suppressed HIV-infected individuals and in an in vitro primary cell model of HIV-1 latency. Our ex vivo and in vitro results demonstrate the association of transcriptional pathways of T cell differentiation, acquisition of effector function, and cell cycle entry in response to LRAs. Analyses of memory cell subsets showed that effector memory pathways and cell surface markers of activation and proliferation in the TEM subset are predictive of higher frequencies of cells carrying an inducible reservoir. Transcriptional profiling also demonstrated that the epigenetic machinery (known to control latency and reactivation) in the TEM subset is associated with frequencies of cells with HIV-integrated DNA and inducible HIV multispliced RNA. TCM cells were triggered to differentiate into TEM cells when they were exposed to LRAs, and this increase of TEM subset frequencies upon LRA stimulation was positively associated with higher numbers of p24+ cells. Together, these data highlight differences in underlying biological latency control in different memory CD4+ T cell subsets which harbor latent HIV in vivo and support a role for differentiation into a TEM phenotype in facilitating latency reversal.IMPORTANCE By performing phenotypic analysis of latency reversal in CD4+ T cells from virally suppressed individuals, we identify the TEM subset as the largest contributor to the inducible HIV reservoir. Differential responses of memory CD4+ T cell subsets to latency-reversing agents (LRAs) demonstrate that HIV gene expression is associated with heightened expression of transcriptional pathways associated with differentiation, acquisition of effector function, and cell cycle entry. In vitro modeling of the latent HIV reservoir in memory CD4+ T cell subsets identify LRAs that reverse latency with ranges of efficiency and specificity. We found that therapeutic induction of latency reversal is associated with upregulation of identical sets of TEM-associated genes and cell surface markers shown to be associated with latency reversal in our ex vivo and in vitro models. Together, these data support the idea that the effector memory phenotype supports HIV latency reversal in CD4+ T cells.
Keywords: CD4 T cells; HIV latency; HIV persistence.
Copyright © 2019 Kulpa et al.
Figures




Similar articles
-
Establishment and Reversal of HIV-1 Latency in Naive and Central Memory CD4+ T Cells In Vitro.J Virol. 2016 Aug 26;90(18):8059-73. doi: 10.1128/JVI.00553-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27356901 Free PMC article.
-
Entry of Polarized Effector Cells into Quiescence Forces HIV Latency.mBio. 2019 Mar 26;10(2):e00337-19. doi: 10.1128/mBio.00337-19. mBio. 2019. PMID: 30914509 Free PMC article.
-
Heterogeneity of Latency Establishment in the Different Human CD4+ T Cell Subsets Stimulated with IL-15.J Virol. 2022 May 25;96(10):e0037922. doi: 10.1128/jvi.00379-22. Epub 2022 May 2. J Virol. 2022. PMID: 35499323 Free PMC article.
-
CD4+ T Cell Subsets and Pathways to HIV Latency.AIDS Res Hum Retroviruses. 2018 Sep;34(9):780-789. doi: 10.1089/AID.2018.0105. Epub 2018 Jul 9. AIDS Res Hum Retroviruses. 2018. PMID: 29869531 Free PMC article. Review.
-
The cell biology of HIV-1 latency and rebound.Retrovirology. 2024 Apr 5;21(1):6. doi: 10.1186/s12977-024-00639-w. Retrovirology. 2024. PMID: 38580979 Free PMC article. Review.
Cited by
-
Dual blockade of IL-10 and PD-1 leads to control of SIV viral rebound following analytical treatment interruption.Nat Immunol. 2024 Oct;25(10):1900-1912. doi: 10.1038/s41590-024-01952-4. Epub 2024 Sep 12. Nat Immunol. 2024. PMID: 39266691 Free PMC article.
-
Spatial technologies to evaluate the HIV-1 reservoir and its microenvironment in the lymph node.mBio. 2024 Aug 14;15(8):e0190924. doi: 10.1128/mbio.01909-24. Epub 2024 Jul 26. mBio. 2024. PMID: 39058091 Free PMC article. Review.
-
A Canadian Survey of Research on HIV-1 Latency-Where Are We Now and Where Are We Heading?Viruses. 2024 Feb 1;16(2):229. doi: 10.3390/v16020229. Viruses. 2024. PMID: 38400005 Free PMC article. Review.
-
HIV Expression in Infected T Cell Clones.Viruses. 2024 Jan 11;16(1):108. doi: 10.3390/v16010108. Viruses. 2024. PMID: 38257808 Free PMC article. Review.
-
Breaking the Silence: Regulation of HIV Transcription and Latency on the Road to a Cure.Viruses. 2023 Dec 15;15(12):2435. doi: 10.3390/v15122435. Viruses. 2023. PMID: 38140676 Free PMC article. Review.
References
-
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi:10.1038/nm.1972. - DOI - PMC - PubMed
-
- Bacchus C, Cheret A, Avettand-Fenoel V, Nembot G, Melard A, Blanc C, Lascoux-Combe C, Slama L, Allegre T, Allavena C, Yazdanpanah Y, Duvivier C, Katlama C, Goujard C, Seksik BC, Leplatois A, Molina JM, Meyer L, Autran B, Rouzioux C, OPTIPRIM ANRS 147 Study Group . 2013. A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PLoS One 8:e64219. doi:10.1371/journal.pone.0064219. - DOI - PMC - PubMed
-
- Yukl SA, Shergill AK, Ho T, Killian M, Girling V, Epling L, Li P, Wong LK, Crouch P, Deeks SG, Havlir DV, McQuaid K, Sinclair E, Wong JK. 2013. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J Infect Dis 208:1212–1220. doi:10.1093/infdis/jit308. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

