Integration of Bioinformatics Resources Reveals the Therapeutic Benefits of Gemcitabine and Cell Cycle Intervention in SMAD4-Deleted Pancreatic Ductal Adenocarcinoma
- PMID: 31569425
- PMCID: PMC6827004
- DOI: 10.3390/genes10100766
Integration of Bioinformatics Resources Reveals the Therapeutic Benefits of Gemcitabine and Cell Cycle Intervention in SMAD4-Deleted Pancreatic Ductal Adenocarcinoma
Abstract
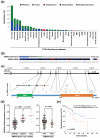
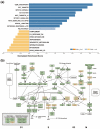
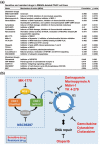
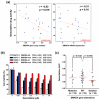
Pancreatic ductal adenocarcinoma (PDAC) is the most common and aggressive type of pancreatic cancer. The five-year survival rate of PDAC is very low (less than 8%), which is associated with the late diagnosis, high metastatic potential, and resistance to therapeutic agents. The identification of better prognostic or therapeutic biomarker may have clinical benefits for PDAC treatment. SMAD4, a central mediator of transforming growth factor beta (TGFβ) signaling pathway, is considered a tumor suppressor gene. SMAD4 inactivation is frequently found in PDAC. However, its role in prognosis and therapeutics of PDAC is still unclear. In this study, we applied bioinformatics approaches, and integrated publicly available resources, to investigate the role of SMAD4 gene deletion in PDAC. We found that SMAD4 deletion was associated with poorer disease-free, but not overall, survival in PDAC patients. Cancer hallmark enrichment and pathway analysis suggested that the upregulation of cell cycle-related genes in SMAD4-deleted PDAC. Chemotherapy response profiling of PDAC cell lines and patient-derived organoids revealed that SMAD4-deleted PDAC was sensitive to gemcitabine, the first-line treatment for PDAC, and specific cell cycle-targeting drugs. Taken together, our study provides an insight into the prognostic and therapeutic roles of SMAD4 gene deletion in PDAC, and SMAD4 gene copy numbers may be used as a therapeutic biomarker for PDAC treatment.
Keywords: SMAD4; bioinformatics; cell cycle; in silico; pancreatic ductal adenocarcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
PDCD10 promotes the tumor-supporting functions of TGF-β in pancreatic cancer.Clin Sci (Lond). 2024 Sep 18;138(18):1111-1129. doi: 10.1042/CS20240450. Clin Sci (Lond). 2024. PMID: 39212293 Free PMC article.
-
Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin signaling in pancreatic ductal adenocarcinoma cells.Gastroenterology. 2014 Aug;147(2):485-97.e18. doi: 10.1053/j.gastro.2014.04.048. Epub 2014 May 20. Gastroenterology. 2014. PMID: 24859161
-
S100A14 promotes progression and gemcitabine resistance in pancreatic cancer.Pancreatology. 2021 Apr;21(3):589-598. doi: 10.1016/j.pan.2021.01.011. Epub 2021 Jan 22. Pancreatology. 2021. PMID: 33579599
-
Localisation of PGK1 determines metabolic phenotype to balance metastasis and proliferation in patients with SMAD4-negative pancreatic cancer.Gut. 2020 May;69(5):888-900. doi: 10.1136/gutjnl-2018-317163. Epub 2019 Oct 14. Gut. 2020. PMID: 31611300 Review.
-
Clinicopathological significance of SMAD4 loss in pancreatic ductal adenocarcinomas: a systematic review and meta-analysis.Oncotarget. 2017 Mar 7;8(10):16704-16711. doi: 10.18632/oncotarget.14335. Oncotarget. 2017. PMID: 28053288 Free PMC article. Review.
Cited by
-
SMAD4 and the TGFβ Pathway in Patients with Pancreatic Ductal Adenocarcinoma.Int J Mol Sci. 2020 May 16;21(10):3534. doi: 10.3390/ijms21103534. Int J Mol Sci. 2020. PMID: 32429474 Free PMC article. Review.
-
Building towards Precision Oncology for Pancreatic Cancer: Real-World Challenges and Opportunities.Genes (Basel). 2020 Sep 21;11(9):1098. doi: 10.3390/genes11091098. Genes (Basel). 2020. PMID: 32967105 Free PMC article. Review.
-
Emerging horizons on molecular and circulating biomarkers in pancreatic adenocarcinoma.Front Oncol. 2024 Nov 7;14:1483306. doi: 10.3389/fonc.2024.1483306. eCollection 2024. Front Oncol. 2024. PMID: 39575418 Free PMC article. Review.
-
KRAS Mutation Variants and Co-occurring PI3K Pathway Alterations Impact Survival for Patients with Pancreatic Ductal Adenocarcinomas.Oncologist. 2022 Dec 9;27(12):1025-1033. doi: 10.1093/oncolo/oyac179. Oncologist. 2022. PMID: 36124727 Free PMC article.
-
Highlights on the Role of KRAS Mutations in Reshaping the Microenvironment of Pancreatic Adenocarcinoma.Int J Mol Sci. 2021 Sep 23;22(19):10219. doi: 10.3390/ijms221910219. Int J Mol Sci. 2021. PMID: 34638560 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

