Isotopically Labeled Clickable Glutathione to Quantify Protein S-Glutathionylation
- PMID: 31560820
- PMCID: PMC7078011
- DOI: 10.1002/cbic.201900528
Isotopically Labeled Clickable Glutathione to Quantify Protein S-Glutathionylation
Abstract
Protein S-glutathionylation is one of the important cysteine oxidation events that regulate various redox-mediated biological processes. Despite several existing methods, there are few proteomic approaches to identify and quantify specific cysteine residues susceptible to S-glutathionylation. We previously developed a clickable glutathione approach that labels intracellular glutathione with azido-Ala by using a mutant form of glutathione synthetase. In this study, we developed a quantification strategy with clickable glutathione by using isotopically labeled heavy and light derivatives of azido-Ala, which provides the relative quantification of glutathionylated peptides in mass spectrometry-based proteomic analysis. We applied isotopically labeled clickable glutathione to HL-1 cardiomyocytes, quantifying relative levels of 1398 glutathionylated peptides upon addition of hydrogen peroxide. Importantly, we highlight elevated levels of glutathionylation on sarcomere-associated muscle proteins while validating glutathionylation of two structural proteins, α-actinin and desmin. Our report provides a chemical proteomic strategy to quantify specific glutathionylated cysteines.
Keywords: S-glutathionylation; clickable glutathione; hydrogen peroxide; protein modification; proteomics; quantification.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest
Figures
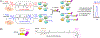
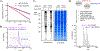

Similar articles
-
Emerging chemistry and biology in protein glutathionylation.Curr Opin Chem Biol. 2022 Dec;71:102221. doi: 10.1016/j.cbpa.2022.102221. Epub 2022 Oct 9. Curr Opin Chem Biol. 2022. PMID: 36223700 Free PMC article. Review.
-
Clickable Glutathione-Based Identification of Cysteine Glutathionylation.Curr Protoc. 2023 Oct;3(10):e907. doi: 10.1002/cpz1.907. Curr Protoc. 2023. PMID: 37818879 Free PMC article.
-
Clickable glutathione using tetrazine-alkene bioorthogonal chemistry for detecting protein glutathionylation.Org Biomol Chem. 2016 Nov 22;14(46):10886-10893. doi: 10.1039/c6ob02050j. Org Biomol Chem. 2016. PMID: 27812596
-
Metabolic synthesis of clickable glutathione for chemoselective detection of glutathionylation.J Am Chem Soc. 2014 Aug 20;136(33):11566-9. doi: 10.1021/ja503946q. Epub 2014 Aug 11. J Am Chem Soc. 2014. PMID: 25079194
-
Assessment of glutathione/glutathione disulphide ratio and S-glutathionylated proteins in human blood, solid tissues, and cultured cells.Free Radic Biol Med. 2017 Nov;112:360-375. doi: 10.1016/j.freeradbiomed.2017.08.008. Epub 2017 Aug 12. Free Radic Biol Med. 2017. PMID: 28807817 Review.
Cited by
-
Contemporary proteomic strategies for cysteine redoxome profiling.Plant Physiol. 2021 May 27;186(1):110-124. doi: 10.1093/plphys/kiaa074. Plant Physiol. 2021. PMID: 33793888 Free PMC article. Review.
-
Chemoproteomic strategy identified p120-catenin glutathionylation regulates E-cadherin degradation and cell migration.Cell Chem Biol. 2023 Dec 21;30(12):1542-1556.e9. doi: 10.1016/j.chembiol.2023.08.004. Epub 2023 Sep 14. Cell Chem Biol. 2023. PMID: 37714153 Free PMC article.
-
Identification and Quantification of Glutathionylated Cysteines under Ischemic Stress.J Proteome Res. 2021 Sep 3;20(9):4529-4542. doi: 10.1021/acs.jproteome.1c00473. Epub 2021 Aug 12. J Proteome Res. 2021. PMID: 34382403 Free PMC article.
-
Emerging chemistry and biology in protein glutathionylation.Curr Opin Chem Biol. 2022 Dec;71:102221. doi: 10.1016/j.cbpa.2022.102221. Epub 2022 Oct 9. Curr Opin Chem Biol. 2022. PMID: 36223700 Free PMC article. Review.
-
Hsp70 in Redox Homeostasis.Cells. 2022 Feb 28;11(5):829. doi: 10.3390/cells11050829. Cells. 2022. PMID: 35269451 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

