Analysis of Sinonasal Microbiota in Exacerbations of Chronic Rhinosinusitis Subgroups
- PMID: 31555757
- PMCID: PMC6749786
- DOI: 10.1177/2473974X19875100
Analysis of Sinonasal Microbiota in Exacerbations of Chronic Rhinosinusitis Subgroups
Abstract
Objective: Microbiome analyses now allow precise determination of the sinus microbiota of patients with exacerbations of chronic rhinosinusitis (CRS). The aim of this report is to describe the sinus microbiota of acute exacerbations in CRS clinical subgroups (with nasal polyps [CRSwNP], without nasal polyps [CRSsNP], and allergic fungal rhinosinusitis [AFRS]).
Study design: Retrospective chart review.
Setting: Tertiary rhinology practice.
Subjects and methods: A retrospective review was performed of all patients whose sinus microbiota were assayed via a commercially available microbiome technology during an acute CRS exacerbation during the 2-year period ending December 31, 2016. All samples were sinus aspirates collected under endoscopic visualization in clinic.
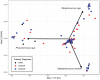
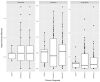
Results: Samples from a total of 134 patients (65 CRSsNP, 55 CRSwNP, and 14 AFRS) were reviewed. The observed richness (number of taxa >2% relative abundance) ranged between 1 and 11 taxa, with an average of 3 taxa per specimen. The most common bacteria in all groups were Staphylococcal spp (including Staphylococcus aureus), Streptococcus spp, Pseudomonas spp, and Escherichia spp. S aureus had an increased prevalence in CRSsNP and AFRS as compared with CRSwNP. Otherwise, the sinus microbiota were markedly similar among all 3 clinical subgroups.
Conclusions: Many bacterial types are identified during acute CRS exacerbation according to DNA-based detection techniques. Bacterial richness was remarkably low in all samples. Few differences in the patterns among clinical subgroups were observed. Further investigation is warranted to determine the clinical significance of these observations and their role in current clinical algorithms.
Keywords: 16S rRNA; bacteria; chronic rhinosinusitis; culture; microbiome; sinusitis.
Figures


Similar articles
-
The Microbiology of Acute Exacerbations in Chronic Rhinosinusitis - A Systematic Review.Front Cell Infect Microbiol. 2022 Mar 24;12:858196. doi: 10.3389/fcimb.2022.858196. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402317 Free PMC article. Review.
-
Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects.Int Forum Allergy Rhinol. 2017 Jun;7(6):561-569. doi: 10.1002/alr.21934. Epub 2017 May 8. Int Forum Allergy Rhinol. 2017. PMID: 28481057
-
Fungal and Bacterial Microbiome in Sinus Mucosa of Patients with and without Chronic Rhinosinusitis.Laryngoscope. 2024 Mar;134(3):1054-1062. doi: 10.1002/lary.30941. Epub 2023 Aug 22. Laryngoscope. 2024. PMID: 37606305
-
The influence of nasal bacterial microbiome diversity on the pathogenesis and prognosis of chronic rhinosinusitis patients with polyps.Eur Arch Otorhinolaryngol. 2021 Apr;278(4):1075-1088. doi: 10.1007/s00405-020-06370-4. Epub 2020 Sep 22. Eur Arch Otorhinolaryngol. 2021. PMID: 32960349
-
Influence of the Microbiome on Chronic Rhinosinusitis With and Without Polyps: An Evolving Discussion.Front Allergy. 2021 Oct 1;2:737086. doi: 10.3389/falgy.2021.737086. eCollection 2021. Front Allergy. 2021. PMID: 35386978 Free PMC article. Review.
Cited by
-
The Microbiology of Acute Exacerbations in Chronic Rhinosinusitis - A Systematic Review.Front Cell Infect Microbiol. 2022 Mar 24;12:858196. doi: 10.3389/fcimb.2022.858196. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402317 Free PMC article. Review.
-
Nasopharyngeal bacterial and fungal microbiota in normal horses and horses with nasopharyngeal cicatrix syndrome.J Vet Intern Med. 2021 Nov;35(6):2897-2911. doi: 10.1111/jvim.16307. Epub 2021 Nov 16. J Vet Intern Med. 2021. PMID: 34783081 Free PMC article.
-
Acute exacerbations of chronic rhinosinusitis: The current state of knowledge.Laryngoscope Investig Otolaryngol. 2022 Jul 20;7(4):935-942. doi: 10.1002/lio2.857. eCollection 2022 Aug. Laryngoscope Investig Otolaryngol. 2022. PMID: 36000029 Free PMC article. Review.
-
Comparison of Fungal and Non-Fungal Rhinosinusitis by Culture-Based Analysis.J Pers Med. 2023 Sep 11;13(9):1368. doi: 10.3390/jpm13091368. J Pers Med. 2023. PMID: 37763136 Free PMC article.
-
The Role of the Gut and Airway Microbiota in Chronic Rhinosinusitis with Nasal Polyps: A Systematic Review.Int J Mol Sci. 2024 Jul 27;25(15):8223. doi: 10.3390/ijms25158223. Int J Mol Sci. 2024. PMID: 39125792 Free PMC article. Review.
References
-
- Ramakrishnan VR, Hauser LJ, Feazel LM, Ir D, Robertson CE, Frank DN. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol. 2015;136:334-342.e1. - PubMed
-
- Psaltis AJ, Wormald PJ. Therapy of sinonasal microbiome in CRS: a critical approach. Curr Allergy Asthma Rep. 2017;17:59. - PubMed
-
- Hauser LJ, Feazel LM, Ir D, et al. Sinus culture poorly predicts resident microbiota. Int Forum Allergy Rhinol. 2015;5:3-9. - PubMed
-
- Jervis Bardy J, Psaltis AJ. Next generation sequencing and the microbiome of chronic rhinosinusitis: a primer for clinicians and review of current research, its limitations, and future directions. Ann Otol Rhinol Laryngol. 2016;125:613-621. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

