Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System
- PMID: 31549070
- PMCID: PMC6750098
- DOI: 10.34133/2019/5641746
Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System
Abstract
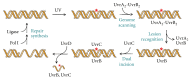
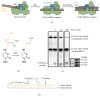
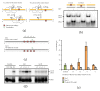
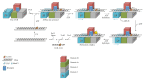
Nucleotide excision repair (NER) is an essential DNA repair system distinguished from other such systems by its extraordinary versatility. NER removes a wide variety of structurally dissimilar lesions having only their bulkiness in common. NER can also repair several less bulky nucleobase lesions, such as 8-oxoguanine. Thus, how a single DNA repair system distinguishes such a diverse array of structurally divergent lesions from undamaged DNA has been one of the great unsolved mysteries in the field of genome maintenance. Here we employ a synthetic crystallography approach to obtain crystal structures of the pivotal NER enzyme UvrB in complex with duplex DNA, trapped at the stage of lesion-recognition. These structures coupled with biochemical studies suggest that UvrB integrates the ATPase-dependent helicase/translocase and lesion-recognition activities. Our work also conclusively establishes the identity of the lesion-containing strand and provides a compelling insight to how UvrB recognizes a diverse array of DNA lesions.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch.Mutat Res. 2000 Aug 30;460(3-4):277-300. doi: 10.1016/s0921-8777(00)00032-x. Mutat Res. 2000. PMID: 10946234 Review.
-
Ribonucleotides as nucleotide excision repair substrates.DNA Repair (Amst). 2014 Jan;13:55-60. doi: 10.1016/j.dnarep.2013.10.010. Epub 2013 Nov 26. DNA Repair (Amst). 2014. PMID: 24290807 Free PMC article.
-
Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.EMBO J. 1999 Dec 15;18(24):6899-907. doi: 10.1093/emboj/18.24.6899. EMBO J. 1999. PMID: 10601012 Free PMC article.
-
Real-time investigation of the roles of ATP hydrolysis by UvrA and UvrB during DNA damage recognition in nucleotide excision repair.DNA Repair (Amst). 2021 Jan;97:103024. doi: 10.1016/j.dnarep.2020.103024. Epub 2020 Nov 25. DNA Repair (Amst). 2021. PMID: 33302090
-
Helicases required for nucleotide excision repair: structure, function and mechanism.Enzymes. 2023;54:273-304. doi: 10.1016/bs.enz.2023.05.002. Epub 2023 Jun 3. Enzymes. 2023. PMID: 37945175 Review.
Cited by
-
A Peek Inside the Machines of Bacterial Nucleotide Excision Repair.Int J Mol Sci. 2021 Jan 19;22(2):952. doi: 10.3390/ijms22020952. Int J Mol Sci. 2021. PMID: 33477956 Free PMC article. Review.
-
Crucial role and mechanism of transcription-coupled DNA repair in bacteria.Nature. 2022 Apr;604(7904):152-159. doi: 10.1038/s41586-022-04530-6. Epub 2022 Mar 30. Nature. 2022. PMID: 35355008 Free PMC article.
-
Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.Nucleic Acids Res. 2023 Apr 11;51(6):2931-2949. doi: 10.1093/nar/gkad108. Nucleic Acids Res. 2023. PMID: 36869664 Free PMC article.
-
Single-molecule studies of helicases and translocases in prokaryotic genome-maintenance pathways.DNA Repair (Amst). 2021 Dec;108:103229. doi: 10.1016/j.dnarep.2021.103229. Epub 2021 Sep 20. DNA Repair (Amst). 2021. PMID: 34601381 Free PMC article. Review.
-
Age-related elevation of O-GlcNAc causes meiotic arrest in male mice.Cell Death Discov. 2023 May 15;9(1):163. doi: 10.1038/s41420-023-01433-x. Cell Death Discov. 2023. PMID: 37188682 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

