Endosomal PI(3)P regulation by the COMMD/CCDC22/CCDC93 (CCC) complex controls membrane protein recycling
- PMID: 31537807
- PMCID: PMC6753146
- DOI: 10.1038/s41467-019-12221-6
Endosomal PI(3)P regulation by the COMMD/CCDC22/CCDC93 (CCC) complex controls membrane protein recycling
Abstract
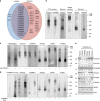
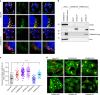
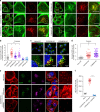
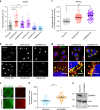
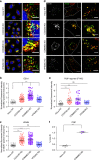
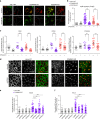
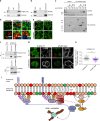
Protein recycling through the endolysosomal system relies on molecular assemblies that interact with cargo proteins, membranes, and effector molecules. Among them, the COMMD/CCDC22/CCDC93 (CCC) complex plays a critical role in recycling events. While CCC is closely associated with retriever, a cargo recognition complex, its mechanism of action remains unexplained. Herein we show that CCC and retriever are closely linked through sharing a common subunit (VPS35L), yet the integrity of CCC, but not retriever, is required to maintain normal endosomal levels of phosphatidylinositol-3-phosphate (PI(3)P). CCC complex depletion leads to elevated PI(3)P levels, enhanced recruitment and activation of WASH (an actin nucleation promoting factor), excess endosomal F-actin and trapping of internalized receptors. Mechanistically, we find that CCC regulates the phosphorylation and endosomal recruitment of the PI(3)P phosphatase MTMR2. Taken together, we show that the regulation of PI(3)P levels by the CCC complex is critical to protein recycling in the endosomal compartment.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A.Mol Biol Cell. 2015 Jan 1;26(1):91-103. doi: 10.1091/mbc.E14-06-1073. Epub 2014 Oct 29. Mol Biol Cell. 2015. PMID: 25355947 Free PMC article.
-
The COMMD Family Regulates Plasma LDL Levels and Attenuates Atherosclerosis Through Stabilizing the CCC Complex in Endosomal LDLR Trafficking.Circ Res. 2018 Jun 8;122(12):1648-1660. doi: 10.1161/CIRCRESAHA.117.312004. Epub 2018 Mar 15. Circ Res. 2018. PMID: 29545368
-
Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling.Nat Cell Biol. 2017 Oct;19(10):1214-1225. doi: 10.1038/ncb3610. Epub 2017 Sep 11. Nat Cell Biol. 2017. PMID: 28892079 Free PMC article.
-
Towards a molecular understanding of endosomal trafficking by Retromer and Retriever.Traffic. 2019 Jul;20(7):465-478. doi: 10.1111/tra.12649. Traffic. 2019. PMID: 30993794 Review.
-
Actin-dependent endosomal receptor recycling.Curr Opin Cell Biol. 2019 Feb;56:22-33. doi: 10.1016/j.ceb.2018.08.006. Epub 2018 Sep 15. Curr Opin Cell Biol. 2019. PMID: 30227382 Review.
Cited by
-
Cell-specific secretory granule sorting mechanisms: the role of MAGEL2 and retromer in hypothalamic regulated secretion.Front Cell Dev Biol. 2023 Sep 18;11:1243038. doi: 10.3389/fcell.2023.1243038. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37799273 Free PMC article. Review.
-
Structural Organization of the Retriever-CCC Endosomal Recycling Complex.bioRxiv [Preprint]. 2023 Jun 7:2023.06.06.543888. doi: 10.1101/2023.06.06.543888. bioRxiv. 2023. Update in: Nat Struct Mol Biol. 2024 Jun;31(6):910-924. doi: 10.1038/s41594-023-01184-4 PMID: 37333304 Free PMC article. Updated. Preprint.
-
Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.Cell. 2023 May 11;186(10):2219-2237.e29. doi: 10.1016/j.cell.2023.04.003. Cell. 2023. PMID: 37172566 Free PMC article.
-
KIF13A-A Key Regulator of Recycling Endosome Dynamics.Front Cell Dev Biol. 2022 Apr 25;10:877532. doi: 10.3389/fcell.2022.877532. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35547822 Free PMC article. Review.
-
FAM21 is critical for TLR2/CLEC4E-mediated dendritic cell function against Candida albicans.Life Sci Alliance. 2023 Jan 30;6(4):e202201414. doi: 10.26508/lsa.202201414. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36717248 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

