A Puzzling Anomaly in the 4-Mer Composition of the Giant Pandoravirus Genomes Reveals a Stringent New Evolutionary Selection Process
- PMID: 31534042
- PMCID: PMC6854483
- DOI: 10.1128/JVI.01206-19
A Puzzling Anomaly in the 4-Mer Composition of the Giant Pandoravirus Genomes Reveals a Stringent New Evolutionary Selection Process
Abstract
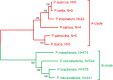
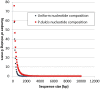
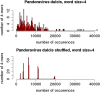
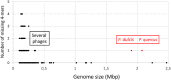
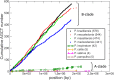
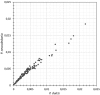
Pandoraviridae is a rapidly growing family of giant viruses, all of which have been isolated using laboratory strains of Acanthamoeba The genomes of 10 distinct strains have been fully characterized, reaching up to 2.5 Mb in size. These double-stranded DNA genomes encode the largest of all known viral proteomes and are propagated in oblate virions that are among the largest ever described (1.2 μm long and 0.5 μm wide). The evolutionary origin of these atypical viruses is the object of numerous speculations. Applying the chaos game representation to the pandoravirus genome sequences, we discovered that the tetranucleotide (4-mer) "AGCT" is totally absent from the genomes of 2 strains (Pandoravirus dulcis and Pandoravirus quercus) and strongly underrepresented in others. Given the amazingly low probability of such an observation in the corresponding randomized sequences, we investigated its biological significance through a comprehensive study of the 4-mer compositions of all viral genomes. Our results indicate that AGCT was specifically eliminated during the evolution of the Pandoraviridae and that none of the previously proposed host-virus antagonistic relationships could explain this phenomenon. Unlike the three other families of giant viruses (Mimiviridae, Pithoviridae, and Molliviridae) infecting the same Acanthamoeba host, the pandoraviruses exhibit a puzzling genomic anomaly suggesting a highly specific DNA editing in response to a new kind of strong evolutionary pressure.IMPORTANCE Recent years have seen the discovery of several families of giant DNA viruses infecting the ubiquitous amoebozoa of the genus Acanthamoeba With double-stranded DNA (dsDNA) genomes reaching 2.5 Mb in length packaged in oblate particles the size of a bacterium, the pandoraviruses are currently the most complex and largest viruses known. In addition to their spectacular dimensions, the pandoraviruses encode the largest proportion of proteins without homologs in other organisms, which is thought to result from a de novo gene creation process. While using comparative genomics to investigate the evolutionary forces responsible for the emergence of such an unusual giant virus family, we discovered a unique bias in the tetranucleotide composition of the pandoravirus genomes that can result only from an undescribed evolutionary process not encountered in any other microorganism.
Keywords: 4-mer statistics; DNA editing; Pandoravirus; chaos game representation; genome composition; giant viruses; host-virus relationship.
Copyright © 2019 American Society for Microbiology.
Figures


Similar articles
-
Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article.
-
New Isolates of Pandoraviruses: Contribution to the Study of Replication Cycle Steps.J Virol. 2019 Feb 19;93(5):e01942-18. doi: 10.1128/JVI.01942-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541841 Free PMC article.
-
Diversity and evolution of the emerging Pandoraviridae family.Nat Commun. 2018 Jun 11;9(1):2285. doi: 10.1038/s41467-018-04698-4. Nat Commun. 2018. PMID: 29891839 Free PMC article.
-
Giant viruses come of age.Curr Opin Microbiol. 2016 Jun;31:50-57. doi: 10.1016/j.mib.2016.03.001. Epub 2016 Mar 19. Curr Opin Microbiol. 2016. PMID: 26999382 Review.
-
Updating strategies for isolating and discovering giant viruses.Curr Opin Microbiol. 2016 Jun;31:80-87. doi: 10.1016/j.mib.2016.03.004. Epub 2016 Apr 1. Curr Opin Microbiol. 2016. PMID: 27039269 Review.
Cited by
-
Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article.
References
-
- Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi:10.1126/science.1239181. - DOI - PubMed
-
- Legendre M, Fabre E, Poirot O, Jeudy S, Lartigue A, Alempic JM, Beucher L, Philippe N, Bertaux L, Christo-Foroux E, Labadie K, Couté Y, Abergel C, Claverie JM. 2018. Diversity and evolution of the emerging Pandoraviridae family. Nat Commun 9:2285. doi:10.1038/s41467-018-04698-4. - DOI - PMC - PubMed
-
- Legendre M, Alempic JM, Philippe N, Lartigue A, Jeudy S, Poirot O, Ta NT, Nin S, Couté Y, Abergel C, Claverie JM. 2019. Pandoravirus celtis illustrates the microevolution processes at work in the giant Pandoraviridae genomes. Front Microbiol 10:430. doi:10.3389/fmicb.2019.00430. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

