Polymer Modeling Predicts Chromosome Reorganization in Senescence
- PMID: 31533042
- PMCID: PMC6859504
- DOI: 10.1016/j.celrep.2019.08.045
Polymer Modeling Predicts Chromosome Reorganization in Senescence
Abstract
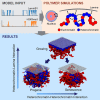
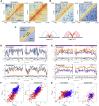
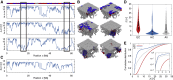
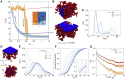
Lamina-associated domains (LADs) cover a large part of the human genome and are thought to play a major role in shaping the nuclear architectural landscape. Here, we perform polymer simulations, microscopy, and mass spectrometry to dissect the roles played by heterochromatin- and lamina-mediated interactions in nuclear organization. Our model explains the conventional organization of heterochromatin and euchromatin in growing cells and the pathological organization found in oncogene-induced senescence and progeria. We show that the experimentally observed changes in the locality of contacts in senescent and progeroid cells can be explained as arising due to phase transitions in the system. Within our simulations, LADs are highly stochastic, as in experiments. Our model suggests that, once established, the senescent phenotype should be metastable even if lamina-mediated interactions were reinstated. Overall, our simulations uncover a generic physical mechanism that can regulate heterochromatin segregation and LAD formation in a wide range of mammalian nuclei.
Keywords: cellular senescence; genome organization; heterochromatin; nuclear lamina; phase transitions; polymer simulations; progeria.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Heterochromatin drives compartmentalization of inverted and conventional nuclei.Nature. 2019 Jun;570(7761):395-399. doi: 10.1038/s41586-019-1275-3. Epub 2019 Jun 5. Nature. 2019. PMID: 31168090 Free PMC article.
-
Lamina-associated domains: peripheral matters and internal affairs.Genome Biol. 2020 Apr 2;21(1):85. doi: 10.1186/s13059-020-02003-5. Genome Biol. 2020. PMID: 32241294 Free PMC article. Review.
-
Restructuring of Lamina-Associated Domains in Senescence and Cancer.Cells. 2022 Jun 5;11(11):1846. doi: 10.3390/cells11111846. Cells. 2022. PMID: 35681541 Free PMC article. Review.
-
Chromatin phase separation and nuclear shape fluctuations are correlated in a polymer model of the nucleus.Nucleus. 2024 Dec;15(1):2351957. doi: 10.1080/19491034.2024.2351957. Epub 2024 May 16. Nucleus. 2024. PMID: 38753956 Free PMC article.
-
The Nuclear Lamina as an Organizer of Chromosome Architecture.Cells. 2019 Feb 8;8(2):136. doi: 10.3390/cells8020136. Cells. 2019. PMID: 30744037 Free PMC article. Review.
Cited by
-
A Lamin-Associated Chromatin Model for Chromosome Organization.Biophys J. 2020 Jun 16;118(12):3041-3050. doi: 10.1016/j.bpj.2020.05.014. Epub 2020 May 20. Biophys J. 2020. PMID: 32492372 Free PMC article.
-
Mechanistic modeling of chromatin folding to understand function.Nat Methods. 2020 Aug;17(8):767-775. doi: 10.1038/s41592-020-0852-6. Epub 2020 Jun 8. Nat Methods. 2020. PMID: 32514111 Review.
-
Mesoscale, long-time mixing of chromosomes and its connection to polymer dynamics.PLoS Comput Biol. 2023 May 25;19(5):e1011142. doi: 10.1371/journal.pcbi.1011142. eCollection 2023 May. PLoS Comput Biol. 2023. PMID: 37228178 Free PMC article.
-
Phase Separation and Correlated Motions in Motorized Genome.J Phys Chem B. 2022 Aug 4;126(30):5619-5628. doi: 10.1021/acs.jpcb.2c03238. Epub 2022 Jul 20. J Phys Chem B. 2022. PMID: 35858189 Free PMC article.
-
Biology and Model Predictions of the Dynamics and Heterogeneity of Chromatin-Nuclear Lamina Interactions.Front Cell Dev Biol. 2022 May 26;10:913458. doi: 10.3389/fcell.2022.913458. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35693945 Free PMC article. Review.
References
-
- Binder K. Theory of first-order phase transitions. Rep. Prog. Phys. 1987;50:783–859.
-
- Bonev B., Cavalli G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016;17:661–678. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

