A Novel Capsid Binding Inhibitor Displays Potent Antiviral Activity against Enterovirus D68
- PMID: 31532189
- PMCID: PMC7167248
- DOI: 10.1021/acsinfecdis.9b00284
A Novel Capsid Binding Inhibitor Displays Potent Antiviral Activity against Enterovirus D68
Abstract
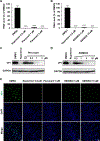
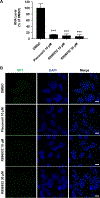
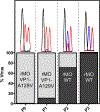
Enterovirus D68 (EV-D68) is a respiratory viral pathogen that primarily infects children under the age of 8. Although EV-D68 infection typically leads to moderate to severe respiratory illnesses, recent years have seen increasing cases of EV-D68 triggered neurological complications such as acute flaccid myelitis (AFM). There is currently no vaccine or antiviral available for EV-D68; we therefore aimed to develop potent and specific small molecule antivirals against EV-D68. In this study, we report our discovery of a viral capsid inhibitor R856932 that inhibits multiple contemporary EV-D68 strains with single-digit to submicromolar efficacy. Mechanistic studies have shown that the tetrazole compound R856932 binds to the hydrophobic pocket of viral capsid protein VP1, thereby preventing viral uncoating and release of viral genome in the infected cells. The mechanism of action of R856932 was confirmed by time-of-addition, Western blot, RT-qPCR, viral heat inactivation, serial viral passage, and reverse genetics experiments. A single mutation located at VP1, A129V, confers resistance against R856932. However, a recombination virus encoding VP1-A129V appeared to have compromised fitness of replication compared to the wild-type EV-D68 virus as shown by the competition growth assay. Overall, the hit compound identified in this study, R856932, represents a promising starting point with a confirmed mechanism of action that can be further developed into EV-D68 antivirals.
Keywords: EV-D68; antiviral; capsid inhibitor; enterovirus; pleconaril.
Figures
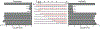

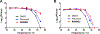
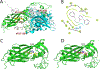

Similar articles
-
Pharmacological Characterization of the Mechanism of Action of R523062, a Promising Antiviral for Enterovirus D68.ACS Infect Dis. 2020 Aug 14;6(8):2260-2270. doi: 10.1021/acsinfecdis.0c00383. Epub 2020 Aug 5. ACS Infect Dis. 2020. PMID: 32692536 Free PMC article.
-
Validating Enterovirus D68-2Apro as an Antiviral Drug Target and the Discovery of Telaprevir as a Potent D68-2Apro Inhibitor.J Virol. 2019 Mar 21;93(7):e02221-18. doi: 10.1128/JVI.02221-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30674624 Free PMC article.
-
Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.Science. 2015 Jan 2;347(6217):71-4. doi: 10.1126/science.1261962. Science. 2015. PMID: 25554786 Free PMC article.
-
Enterovirus D68 Antivirals: Past, Present, and Future.ACS Infect Dis. 2020 Jul 10;6(7):1572-1586. doi: 10.1021/acsinfecdis.0c00120. Epub 2020 May 14. ACS Infect Dis. 2020. PMID: 32352280 Free PMC article. Review.
-
Regulation of the proteostasis network during enterovirus infection: A feedforward mechanism for EV-A71 and EV-D68.Antiviral Res. 2021 Apr;188:105019. doi: 10.1016/j.antiviral.2021.105019. Epub 2021 Jan 20. Antiviral Res. 2021. PMID: 33484748 Review.
Cited by
-
Discovery of Potent and Broad-Spectrum Pyrazolopyridine-Containing Antivirals against Enteroviruses D68, A71, and Coxsackievirus B3 by Targeting the Viral 2C Protein.J Med Chem. 2021 Jun 24;64(12):8755-8774. doi: 10.1021/acs.jmedchem.1c00758. Epub 2021 Jun 4. J Med Chem. 2021. PMID: 34085827 Free PMC article.
-
Pharmacological Characterization of the Mechanism of Action of R523062, a Promising Antiviral for Enterovirus D68.ACS Infect Dis. 2020 Aug 14;6(8):2260-2270. doi: 10.1021/acsinfecdis.0c00383. Epub 2020 Aug 5. ACS Infect Dis. 2020. PMID: 32692536 Free PMC article.
-
Enterovirus A71 antivirals: Past, present, and future.Acta Pharm Sin B. 2022 Apr;12(4):1542-1566. doi: 10.1016/j.apsb.2021.08.017. Epub 2021 Aug 20. Acta Pharm Sin B. 2022. PMID: 35847514 Free PMC article.
-
Enterovirus D68 molecular and cellular biology and pathogenesis.J Biol Chem. 2021 Jan-Jun;296:100317. doi: 10.1016/j.jbc.2021.100317. Epub 2021 Jan 21. J Biol Chem. 2021. PMID: 33484714 Free PMC article. Review.
-
Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors.ACS Pharmacol Transl Sci. 2020 Oct 9;3(6):1265-1277. doi: 10.1021/acsptsci.0c00130. eCollection 2020 Dec 11. ACS Pharmacol Transl Sci. 2020. PMID: 33330841 Free PMC article.
References
-
- Baggen J; Thibaut HJ; Strating JRPM; van Kuppeveld FJM The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol 2018, 16, 368–381. - PubMed
-
- Royston L; Essaidi-Laziosi M; Perez-Rodriguez FJ; Piuz I; Geiser J; Krause KH; Huang S; Constant S; Kaiser L; Garcin D; Tapparel C Viral chimeras decrypt the role of enterovirus capsid proteins in viral tropism, acid sensitivity and optimal growth temperature. PLoS Pathog 2018, 14, e1006962. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

