RCSB Protein Data Bank: Enabling biomedical research and drug discovery
- PMID: 31531901
- PMCID: PMC6933845
- DOI: 10.1002/pro.3730
RCSB Protein Data Bank: Enabling biomedical research and drug discovery
Abstract
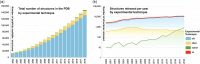
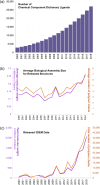
Analyses of publicly available structural data reveal interesting insights into the impact of the three-dimensional (3D) structures of protein targets important for discovery of new drugs (e.g., G-protein-coupled receptors, voltage-gated ion channels, ligand-gated ion channels, transporters, and E3 ubiquitin ligases). The Protein Data Bank (PDB) archive currently holds > 155,000 atomic-level 3D structures of biomolecules experimentally determined using crystallography, nuclear magnetic resonance spectroscopy, and electron microscopy. The PDB was established in 1971 as the first open-access, digital-data resource in biology, and is now managed by the Worldwide PDB partnership (wwPDB; wwPDB.org). US PDB operations are the responsibility of the Research Collaboratory for Structural Bioinformatics PDB (RCSB PDB). The RCSB PDB serves millions of RCSB.org users worldwide by delivering PDB data integrated with ∼40 external biodata resources, providing rich structural views of fundamental biology, biomedicine, and energy sciences. Recently published work showed that the PDB archival holdings facilitated discovery of ∼90% of the 210 new drugs approved by the US Food and Drug Administration 2010-2016. We review user-driven development of RCSB PDB services, examine growth of the PDB archive in terms of size and complexity, and present examples and opportunities for structure-guided drug discovery for challenging targets (e.g., integral membrane proteins).
Keywords: GPCR; Protein Data Bank; integral membrane proteins; ion channel; protein structure and function; structural biology; structure-guided drug discovery; transporter; ubiquitin ligase.
© 2019 The Protein Society.
Figures


Similar articles
-
RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education.Protein Sci. 2018 Jan;27(1):316-330. doi: 10.1002/pro.3331. Epub 2017 Nov 11. Protein Sci. 2018. PMID: 29067736 Free PMC article. Review.
-
RCSB Protein Data bank: Tools for visualizing and understanding biological macromolecules in 3D.Protein Sci. 2022 Dec;31(12):e4482. doi: 10.1002/pro.4482. Protein Sci. 2022. PMID: 36281733 Free PMC article.
-
RCSB Protein Data Bank: supporting research and education worldwide through explorations of experimentally determined and computationally predicted atomic level 3D biostructures.IUCrJ. 2024 May 1;11(Pt 3):279-286. doi: 10.1107/S2052252524002604. IUCrJ. 2024. PMID: 38597878 Free PMC article.
-
RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences.Nucleic Acids Res. 2021 Jan 8;49(D1):D437-D451. doi: 10.1093/nar/gkaa1038. Nucleic Acids Res. 2021. PMID: 33211854 Free PMC article.
-
Protein Data Bank: A Comprehensive Review of 3D Structure Holdings and Worldwide Utilization by Researchers, Educators, and Students.Biomolecules. 2022 Oct 4;12(10):1425. doi: 10.3390/biom12101425. Biomolecules. 2022. PMID: 36291635 Free PMC article. Review.
Cited by
-
Computational drug repurposing study elucidating simultaneous inhibition of entry and replication of novel corona virus by Grazoprevir.Sci Rep. 2021 Mar 31;11(1):7307. doi: 10.1038/s41598-021-86712-2. Sci Rep. 2021. PMID: 33790352 Free PMC article.
-
Recent Developments in New Therapeutic Agents against Alzheimer and Parkinson Diseases: In-Silico Approaches.Molecules. 2021 Apr 11;26(8):2193. doi: 10.3390/molecules26082193. Molecules. 2021. PMID: 33920326 Free PMC article. Review.
-
GPCR-BERT: Interpreting Sequential Design of G Protein-Coupled Receptors Using Protein Language Models.J Chem Inf Model. 2024 Feb 26;64(4):1134-1144. doi: 10.1021/acs.jcim.3c01706. Epub 2024 Feb 10. J Chem Inf Model. 2024. PMID: 38340054 Free PMC article.
-
The natural breakthrough: phytochemicals as potent therapeutic agents against spinocerebellar ataxia type 3.Sci Rep. 2024 Jan 17;14(1):1529. doi: 10.1038/s41598-024-51954-3. Sci Rep. 2024. PMID: 38233440 Free PMC article.
-
PDBe CCDUtils: an RDKit-based toolkit for handling and analysing small molecules in the Protein Data Bank.J Cheminform. 2023 Dec 2;15(1):117. doi: 10.1186/s13321-023-00786-w. J Cheminform. 2023. PMID: 38042830 Free PMC article.
References
-
- Protein Data Bank . Protein Data Bank. Nature New Biol. 1971;233:223. - PubMed
-
- Protein Data Bank . Cold spring harbor symposia on quantitative biology. Vol 36 New York, NY: Cold Spring Laboratory Press, 1972.
-
- Sullivan KP, Brennan‐Tonetta P, Marxen LJ (2017) Economic impacts of the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank. doi: 10.2210/rcsb_pdb/pdb-econ-imp-2017. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

