Trim32 suppresses cerebellar development and tumorigenesis by degrading Gli1/sonic hedgehog signaling
- PMID: 31527798
- PMCID: PMC7206143
- DOI: 10.1038/s41418-019-0415-5
Trim32 suppresses cerebellar development and tumorigenesis by degrading Gli1/sonic hedgehog signaling
Abstract
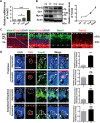
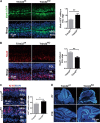
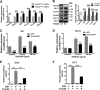
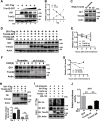
Sonic hedgehog (SHH) signaling is crucial for the maintenance of the physiological self-renewal of granule neuron progenitor cells (GNPs) during cerebellar development, and its dysregulation leads to oncogenesis. However, how SHH signaling is controlled during cerebellar development is poorly understood. Here, we show that Trim32, a cell fate determinant, is distributed asymmetrically in the cytoplasm of mitotic GNPs, and that genetic knockout of Trim32 keeps GNPs at a proliferating and undifferentiated state. In addition, Trim32 knockout enhances the incidence of medulloblastoma (MB) formation in the Ptch1 mutant mice. Mechanistically, Trim32 binds to Gli1, an effector of SHH signaling, via its NHL domain and degrades the latter through its RING domain to antagonize the SHH pathway. These findings provide a novel mechanism that Trim32 may be a vital cell fate regulator by antagonizing the SHH signaling to promote GNPs differentiation and a tumor suppressor in MB formation.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures

Similar articles
-
The p53 inhibitor MDM2 facilitates Sonic Hedgehog-mediated tumorigenesis and influences cerebellar foliation.PLoS One. 2011 Mar 18;6(3):e17884. doi: 10.1371/journal.pone.0017884. PLoS One. 2011. PMID: 21437245 Free PMC article.
-
RNF220 is required for cerebellum development and regulates medulloblastoma progression through epigenetic modulation of Shh signaling.Development. 2020 Jun 15;147(21):dev188078. doi: 10.1242/dev.188078. Development. 2020. PMID: 32376680
-
Astrocytes Promote Medulloblastoma Progression through Hedgehog Secretion.Cancer Res. 2017 Dec 1;77(23):6692-6703. doi: 10.1158/0008-5472.CAN-17-1463. Epub 2017 Oct 6. Cancer Res. 2017. PMID: 28986380 Free PMC article.
-
Hedgehog Signaling and Truncated GLI1 in Cancer.Cells. 2020 Sep 17;9(9):2114. doi: 10.3390/cells9092114. Cells. 2020. PMID: 32957513 Free PMC article. Review.
-
TRIM32: A Multifunctional Protein Involved in Muscle Homeostasis, Glucose Metabolism, and Tumorigenesis.Biomolecules. 2021 Mar 10;11(3):408. doi: 10.3390/biom11030408. Biomolecules. 2021. PMID: 33802079 Free PMC article. Review.
Cited by
-
Common germline-somatic variant interactions in advanced urothelial cancer.Nat Commun. 2020 Dec 3;11(1):6195. doi: 10.1038/s41467-020-19971-8. Nat Commun. 2020. PMID: 33273457 Free PMC article.
-
Drosophila p53 tumor suppressor directly activates conserved asymmetric stem cell division regulators.iScience. 2024 Oct 9;27(11):111118. doi: 10.1016/j.isci.2024.111118. eCollection 2024 Nov 15. iScience. 2024. PMID: 39524346 Free PMC article.
-
Deficiency of TRIM32 Impairs Motor Function and Purkinje Cells in Mid-Aged Mice.Front Aging Neurosci. 2021 Aug 6;13:697494. doi: 10.3389/fnagi.2021.697494. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34421574 Free PMC article.
-
CDK2-activated TRIM32 phosphorylation and nuclear translocation promotes radioresistance in triple-negative breast cancer.J Adv Res. 2024 Jul;61:239-251. doi: 10.1016/j.jare.2023.09.011. Epub 2023 Sep 19. J Adv Res. 2024. PMID: 37734566 Free PMC article.
-
Tripartite Motif-Containing Protein 32 (TRIM32): What Does It Do for Skeletal Muscle?Cells. 2023 Aug 19;12(16):2104. doi: 10.3390/cells12162104. Cells. 2023. PMID: 37626915 Free PMC article. Review.
References
-
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

