Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPα and canonical Wnt signaling
- PMID: 31515577
- PMCID: PMC11105029
- DOI: 10.1007/s00018-019-03289-w
Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPα and canonical Wnt signaling
Abstract
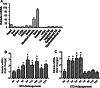
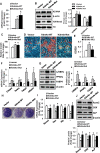
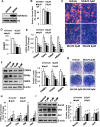
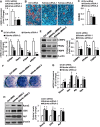
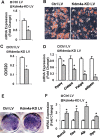
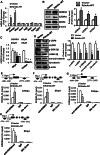
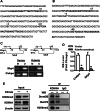
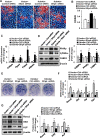
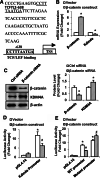
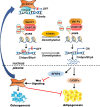
Epigenetic modifications play a central role in cell differentiation and development. In the current study, we have recognized lysine demethylase 4A (KDM4A) as a novel epigenetic regulator of osteoblast and adipocyte differentiation. Kdm4a expression was upregulated during osteogenesis and adipogenesis of primary marrow stromal cells and established stromal ST2 line. Overexpression of wild-type Kdm4a promoted adipogenic differentiation and blocked osteogenic differentiation of the progenitor cells. This effect was largely alleviated when the catalytically dead mutation was made. Conversely, depletion or inactivation of Kdm4a in undifferentiated progenitor cells inhibited the formation of adipocytes and promoted the differentiation of osteoblasts. Mechanism explorations showed that overexpression of Kdm4a upregulated the expression of secreted frizzled-related protein 4 (Sfrp4) and CCAAT/enhancer-binding protein α (C/ebpα). Chromatin immunoprecipitation assay demonstrated that KDM4A directly bound the promoters of Sfrp4 and C/ebpα, removed the histone methylation mark H3K9me3, and reduced DNA methylation levels of CpG in promoter regions of C/ebpα and Sfrp4. Furthermore, overexpression of Kdm4a inactivated canonical Wnt signaling. Moreover, activation of canonical Wnt signaling through silencing of Sfrp4 in ST2 attenuated the inhibition of osteogenic differentiation and the enhancement of adipogenic differentiation by KDM4A. These data have identified KDM4A as a novel regulator of osteoblast and adipocyte differentiation and suggest KDM4A inhibition as a potential therapeutic target for treating metabolic disorders such as osteoporosis.
Keywords: Adipocyte; CCAAT/enhancer-binding protein α; Differentiation; Lysine demethylase 4A; Osteoblast; Secreted frizzled-related protein 4; Wnt/β-catenin.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures
Similar articles
-
Histone demethylase KDM7A reciprocally regulates adipogenic and osteogenic differentiation via regulation of C/EBPα and canonical Wnt signalling.J Cell Mol Med. 2019 Mar;23(3):2149-2162. doi: 10.1111/jcmm.14126. Epub 2019 Jan 7. J Cell Mol Med. 2019. PMID: 30614617 Free PMC article.
-
Nuclear factor I-C reciprocally regulates adipocyte and osteoblast differentiation via control of canonical Wnt signaling.FASEB J. 2017 May;31(5):1939-1952. doi: 10.1096/fj.201600975RR. Epub 2017 Jan 25. FASEB J. 2017. PMID: 28122918
-
A novel Sprouty4-ERK1/2-Wnt/β-catenin regulatory loop in marrow stromal progenitor cells controls osteogenic and adipogenic differentiation.Metabolism. 2020 Apr;105:154189. doi: 10.1016/j.metabol.2020.154189. Epub 2020 Feb 24. Metabolism. 2020. PMID: 32105664
-
PPARγ and Wnt Signaling in Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells.Curr Stem Cell Res Ther. 2016;11(3):216-25. doi: 10.2174/1574888x10666150519093429. Curr Stem Cell Res Ther. 2016. PMID: 25986621 Review.
-
Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells.Int J Oral Sci. 2015 Dec 18;7(4):197-204. doi: 10.1038/ijos.2015.41. Int J Oral Sci. 2015. PMID: 26674421 Free PMC article. Review.
Cited by
-
Role of histone modification in the occurrence and development of osteoporosis.Front Endocrinol (Lausanne). 2022 Aug 26;13:964103. doi: 10.3389/fendo.2022.964103. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36093077 Free PMC article. Review.
-
miR‑30a‑5p induces the adipogenic differentiation of bone marrow mesenchymal stem cells by targeting FAM13A/Wnt/β‑catenin signaling in aplastic anemia.Mol Med Rep. 2022 Jan;25(1):27. doi: 10.3892/mmr.2021.12543. Epub 2021 Nov 25. Mol Med Rep. 2022. PMID: 34821370 Free PMC article.
-
Epigenetic Regulation of Autophagy in Bone Metabolism.Function (Oxf). 2024 Jan 27;5(2):zqae004. doi: 10.1093/function/zqae004. eCollection 2024. Function (Oxf). 2024. PMID: 38486976 Free PMC article. Review.
-
The Roles of Epigenetics Regulation in Bone Metabolism and Osteoporosis.Front Cell Dev Biol. 2021 Jan 25;8:619301. doi: 10.3389/fcell.2020.619301. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33569383 Free PMC article. Review.
-
[The Role of Histone Demethylase in Osteogenic and Chondrogenic Differentiation of Mesenchymal Stem Cells: A Literature Review].Sichuan Da Xue Xue Bao Yi Xue Ban. 2021 May;52(3):364-372. doi: 10.12182/20210560202. Sichuan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 34018352 Free PMC article. Review. Chinese.
References
-
- Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T, Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2(3):165–171. - PubMed
-
- Sottile V, Seuwen K. Bone morphogenetic protein-2 stimulates adipogenic differentiation of mesenchymal precursor cells in synergy with BRL 49653 (rosiglitazone) FEBS Lett. 2000;475(3):201–204. - PubMed
MeSH terms
Substances
Grants and funding
- 81871741/National Natural Science Foundation of China
- 81672116/National Natural Science Foundation of China
- 81772297/National Natural Science Foundation of China
- 18JCZDJC32200/Natural Science Foundation of Tianjin Municipal Science and Technology Commission
- 18JCQNJC12900/Natural Science Foundation of Tianjin Municipal Science and Technology Commission
LinkOut - more resources
Full Text Sources

