Functional analysis of Cwc24 ZF-domain in 5' splice site selection
- PMID: 31504764
- PMCID: PMC6821175
- DOI: 10.1093/nar/gkz733
Functional analysis of Cwc24 ZF-domain in 5' splice site selection
Abstract
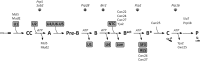
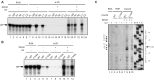
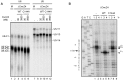
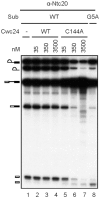
The essential splicing factor Cwc24 contains a zinc-finger (ZF) domain required for its function in splicing. Cwc24 binds over the 5' splice site after the spliceosome is activated, and its binding prior to Prp2-mediated spliceosome remodeling is important for proper interactions of U5 and U6 with the 5' splice site sequence and selection of the 5' splice site. Here, we show that Cwc24 transiently interacts with the 5' splice site in formation of the functional RNA catalytic core during spliceosome remodeling, and the ZF-motif is required for specific interaction of Cwc24 with the 5' splice site. Deletion of the ZF domain or mutation of the conserved ZF residues greatly weakened the association of Cwc24 with the spliceosome, and lowered the affinity and specificity of its interaction with the 5' splice site, resulting in atypical interactions of U5, U6 and Prp8 with the 5' splice site, and aberrant cleavage at the 5' splice site. Our results reveal a crucial role of the Cwc24 ZF-motif for defining 5' splice site selection in the first splicing step.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Role of Cwc24 in the First Catalytic Step of Splicing and Fidelity of 5' Splice Site Selection.Mol Cell Biol. 2017 Mar 1;37(6):e00580-16. doi: 10.1128/MCB.00580-16. Print 2017 Mar 15. Mol Cell Biol. 2017. PMID: 27994011 Free PMC article.
-
Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3' Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes.PLoS Genet. 2015 Sep 22;11(9):e1005539. doi: 10.1371/journal.pgen.1005539. eCollection 2015. PLoS Genet. 2015. PMID: 26393790 Free PMC article.
-
How Is Precursor Messenger RNA Spliced by the Spliceosome?Annu Rev Biochem. 2020 Jun 20;89:333-358. doi: 10.1146/annurev-biochem-013118-111024. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815536 Review.
-
The splicing factor Prp17 interacts with the U2, U5 and U6 snRNPs and associates with the spliceosome pre- and post-catalysis.Biochem J. 2008 Dec 15;416(3):365-74. doi: 10.1042/BJ20081195. Biochem J. 2008. PMID: 18691155
-
The role of PRP8 protein in nuclear pre-mRNA splicing in yeast.J Cell Sci Suppl. 1995;19:101-5. doi: 10.1242/jcs.1995.supplement_19.15. J Cell Sci Suppl. 1995. PMID: 8655640 Review.
Cited by
-
Arresting Spliceosome Intermediates at Various Stages of the Splicing Pathway.Methods Mol Biol. 2023;2666:193-211. doi: 10.1007/978-1-0716-3191-1_15. Methods Mol Biol. 2023. PMID: 37166667
References
-
- Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002; 36:333–360. - PubMed
-
- Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. - PubMed
-
- Will C.L., Lührmann R.. Atkins JF, Gesteland RF, Cech TR. Cold Spring Harb. Perspect. Biol. 2011; 3:NY: Cold Spring Harbor Laboratory Press; a003707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

