'Candidatus Phytoplasma solani' interferes with the distribution and uptake of iron in tomato
- PMID: 31500568
- PMCID: PMC6734453
- DOI: 10.1186/s12864-019-6062-x
'Candidatus Phytoplasma solani' interferes with the distribution and uptake of iron in tomato
Abstract
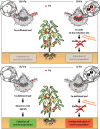
Background: 'Candidatus Phytoplasma solani' is endemic in Europe and infects a wide range of weeds and cultivated plants. Phytoplasmas are prokaryotic plant pathogens that colonize the sieve elements of their host plant, causing severe alterations in phloem function and impairment of assimilate translocation. Typical symptoms of infected plants include yellowing of leaves or shoots, leaf curling, and general stunting, but the molecular mechanisms underlying most of the reported changes remain largely enigmatic. To infer a possible involvement of Fe in the host-phytoplasma interaction, we investigated the effects of 'Candidatus Phytoplasma solani' infection on tomato plants (Solanum lycopersicum cv. Micro-Tom) grown under different Fe regimes.
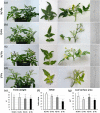
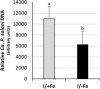
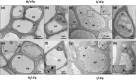
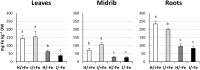
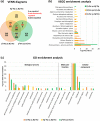
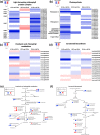
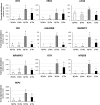
Results: Both phytoplasma infection and Fe starvation led to the development of chlorotic leaves and altered thylakoid organization. In infected plants, Fe accumulated in phloem tissue, altering the local distribution of Fe. In infected plants, Fe starvation had additive effects on chlorophyll content and leaf chlorosis, suggesting that the two conditions affected the phenotypic readout via separate routes. To gain insights into the transcriptional response to phytoplasma infection, or Fe deficiency, transcriptome profiling was performed on midrib-enriched leaves. RNA-seq analysis revealed that both stress conditions altered the expression of a large (> 800) subset of common genes involved in photosynthetic light reactions, porphyrin / chlorophyll metabolism, and in flowering control. In Fe-deficient plants, phytoplasma infection perturbed the Fe deficiency response in roots, possibly by interference with the synthesis or transport of a promotive signal transmitted from the leaves to the roots.
Conclusions: 'Candidatus Phytoplasma solani' infection changes the Fe distribution in tomato leaves, affects the photosynthetic machinery and perturbs the orchestration of root-mediated transport processes by compromising shoot-to-root communication.
Keywords: Carotenoids metabolism; Chlorophyll; Iron deficiency; Leaves; NGS; Phloem; Phytoplasma; Porphyrin; Roots; Tomato.
Conflict of interest statement
The authors declare that they have no competing interets.
Figures

Similar articles
-
Infection by phloem-limited phytoplasma affects mineral nutrient homeostasis in tomato leaf tissues.J Plant Physiol. 2022 Apr;271:153659. doi: 10.1016/j.jplph.2022.153659. Epub 2022 Mar 1. J Plant Physiol. 2022. PMID: 35299031
-
Combined microscopy and molecular analyses show phloem occlusions and cell wall modifications in tomato leaves in response to 'Candidatus Phytoplasma solani'.J Microsc. 2016 Aug;263(2):212-25. doi: 10.1111/jmi.12426. Epub 2016 May 20. J Microsc. 2016. PMID: 27197728
-
Involvement of SUT1 and SUT2 Sugar Transporters in the Impairment of Sugar Transport and Changes in Phloem Exudate Contents in Phytoplasma-Infected Plants.Int J Mol Sci. 2021 Jan 13;22(2):745. doi: 10.3390/ijms22020745. Int J Mol Sci. 2021. PMID: 33451049 Free PMC article.
-
Iron transport mechanisms and their evolution focusing on chloroplasts.J Plant Physiol. 2023 Sep;288:154059. doi: 10.1016/j.jplph.2023.154059. Epub 2023 Jul 31. J Plant Physiol. 2023. PMID: 37586271 Review.
-
Bois noir management in vineyard: a review on effective and promising control strategies.Front Plant Sci. 2024 Mar 27;15:1364241. doi: 10.3389/fpls.2024.1364241. eCollection 2024. Front Plant Sci. 2024. PMID: 38601314 Free PMC article. Review.
Cited by
-
Study of genetic modifications of flower development and methylation status in phytoplasma infected Brassica (Brassica rapa L.).Mol Biol Rep. 2022 Dec;49(12):11359-11369. doi: 10.1007/s11033-022-07743-0. Epub 2022 Aug 2. Mol Biol Rep. 2022. PMID: 35916993
-
Modifying Anthocyanins Biosynthesis in Tomato Hairy Roots: A Test Bed for Plant Resistance to Ionizing Radiation and Antioxidant Properties in Space.Front Plant Sci. 2022 Feb 24;13:830931. doi: 10.3389/fpls.2022.830931. eCollection 2022. Front Plant Sci. 2022. PMID: 35283922 Free PMC article.
-
The sieve-element endoplasmic reticulum: A focal point of phytoplasma-host plant interaction?Front Microbiol. 2023 Feb 2;14:1030414. doi: 10.3389/fmicb.2023.1030414. eCollection 2023. Front Microbiol. 2023. PMID: 36819061 Free PMC article.
-
A newly identified glycosyltransferase AsRCOM provides resistance to purple curl leaf disease in agave.BMC Genomics. 2023 Nov 7;24(1):669. doi: 10.1186/s12864-023-09700-y. BMC Genomics. 2023. PMID: 37936069 Free PMC article.
-
Nano and chelated iron fertilization influences marketable yield, phytochemical properties, and antioxidant capacity of tomatoes.PLoS One. 2023 Nov 8;18(11):e0294033. doi: 10.1371/journal.pone.0294033. eCollection 2023. PLoS One. 2023. PMID: 37939150 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

