Androgen-regulated transcription of ESRP2 drives alternative splicing patterns in prostate cancer
- PMID: 31478829
- PMCID: PMC6788855
- DOI: 10.7554/eLife.47678
Androgen-regulated transcription of ESRP2 drives alternative splicing patterns in prostate cancer
Abstract
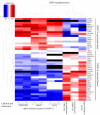
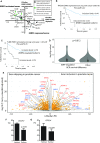
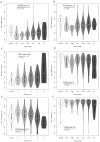
Prostate is the most frequent cancer in men. Prostate cancer progression is driven by androgen steroid hormones, and delayed by androgen deprivation therapy (ADT). Androgens control transcription by stimulating androgen receptor (AR) activity, yet also control pre-mRNA splicing through less clear mechanisms. Here we find androgens regulate splicing through AR-mediated transcriptional control of the epithelial-specific splicing regulator ESRP2. Both ESRP2 and its close paralog ESRP1 are highly expressed in primary prostate cancer. Androgen stimulation induces splicing switches in many endogenous ESRP2-controlled mRNA isoforms, including splicing switches correlating with disease progression. ESRP2 expression in clinical prostate cancer is repressed by ADT, which may thus inadvertently dampen epithelial splice programmes. Supporting this, treatment with the AR antagonist bicalutamide (Casodex) induced mesenchymal splicing patterns of genes including FLNB and CTNND1. Our data reveals a new mechanism of splicing control in prostate cancer with important implications for disease progression.
Keywords: RNA; cancer; chromosomes; gene expression; human; splicing.
© 2019, Munkley et al.
Conflict of interest statement
JM, LL, SK, GH, ES, CD, HO, TM, KC, IE, KL, HZ, OT, BK, PM, JM, MC, LH, MD, SC, NB, SO, DE No competing interests declared
Figures
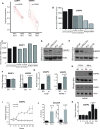
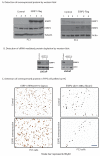
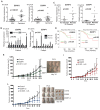

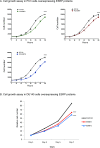

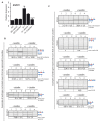

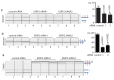
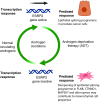
Similar articles
-
Androgen receptor signaling regulates the transcriptome of prostate cancer cells by modulating global alternative splicing.Oncogene. 2020 Sep;39(39):6172-6189. doi: 10.1038/s41388-020-01429-2. Epub 2020 Aug 20. Oncogene. 2020. PMID: 32820253 Free PMC article.
-
Mechanisms of the androgen receptor splicing in prostate cancer cells.Oncogene. 2014 Jun 12;33(24):3140-50. doi: 10.1038/onc.2013.284. Epub 2013 Jul 15. Oncogene. 2014. PMID: 23851510 Free PMC article.
-
The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor.J Pathol. 2008 May;215(1):67-77. doi: 10.1002/path.2324. J Pathol. 2008. PMID: 18273831
-
RNA splicing and splicing regulator changes in prostate cancer pathology.Hum Genet. 2017 Sep;136(9):1143-1154. doi: 10.1007/s00439-017-1792-9. Epub 2017 Apr 5. Hum Genet. 2017. PMID: 28382513 Free PMC article. Review.
-
Androgen receptors in hormone-dependent and castration-resistant prostate cancer.Pharmacol Ther. 2013 Dec;140(3):223-38. doi: 10.1016/j.pharmthera.2013.07.003. Epub 2013 Jul 13. Pharmacol Ther. 2013. PMID: 23859952 Review.
Cited by
-
Role of epithelial splicing regulatory protein 1 in cancer progression.Cancer Cell Int. 2023 Dec 18;23(1):331. doi: 10.1186/s12935-023-03180-6. Cancer Cell Int. 2023. PMID: 38110955 Free PMC article. Review.
-
Sialylation Inhibition Can Partially Revert Acquired Resistance to Enzalutamide in Prostate Cancer Cells.Cancers (Basel). 2024 Aug 24;16(17):2953. doi: 10.3390/cancers16172953. Cancers (Basel). 2024. PMID: 39272811 Free PMC article.
-
The epithelial splicing regulator ESRP2 is epigenetically repressed by DNA hypermethylation in Wilms tumour and acts as a tumour suppressor.Mol Oncol. 2022 Feb;16(3):630-647. doi: 10.1002/1878-0261.13101. Epub 2021 Sep 28. Mol Oncol. 2022. PMID: 34520622 Free PMC article.
-
Alternative splicing in prostate cancer progression and therapeutic resistance.Oncogene. 2024 May;43(22):1655-1668. doi: 10.1038/s41388-024-03036-x. Epub 2024 Apr 24. Oncogene. 2024. PMID: 38658776 Free PMC article. Review.
-
The crosstalk between alternative splicing and circular RNA in cancer: pathogenic insights and therapeutic implications.Cell Mol Biol Lett. 2024 Nov 16;29(1):142. doi: 10.1186/s11658-024-00662-x. Cell Mol Biol Lett. 2024. PMID: 39550559 Free PMC article. Review.
References
-
- Adler D, Kelly ST. R Package Version 0.3.2Vioplot: Violin Plot. 2018 https://github.com/TomKellyGenetics/vioplot
-
- Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, Tse C, Wang K, Mo F, Haegert A, Brahmbhatt S, Bell R, Adomat H, Kawai Y, Xue H, Dong X, Fazli L, Tsai H, Lotan TL, Kossai M, Mosquera JM, Rubin MA, Beltran H, Zoubeidi A, Wang Y, Gleave ME, Collins CC. The placental gene PEG10 promotes progression of neuroendocrine prostate Cancer. Cell Reports. 2015;12:922–936. doi: 10.1016/j.celrep.2015.07.012. - DOI - PubMed
-
- Arredouani MS, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, Sanda MG. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate Cancer. Clinical Cancer Research. 2009;15:5794–5802. doi: 10.1158/1078-0432.CCR-09-0911. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

