Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations
- PMID: 31450634
- PMCID: PMC6769881
- DOI: 10.3390/genes10090642
Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations
Abstract
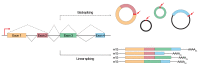
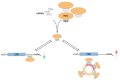
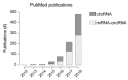
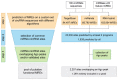
Circular RNAs (circRNAs) have recently emerged as a novel class of transcripts, characterized by covalently linked 3'-5' ends that result in the so-called backsplice junction. During the last few years, thousands of circRNAs have been identified in different organisms. Yet, despite their role as disease biomarker started to emerge, depicting their function remains challenging. Different studies have shown that certain circRNAs act as miRNA sponges, but any attempt to generalize from the single case to the "circ-ome" has failed so far. In this review, we explore the potential to define miRNA "sponging" as a more general function of circRNAs and describe the different approaches to predict miRNA response elements (MREs) in known or novel circRNA sequences. Moreover, we discuss how experiments based on Ago2-IP and experimentally validated miRNA:target duplexes can be used to either prioritize or validate putative miRNA-circRNA associations.
Keywords: circRNA; miRNA; miRNA sponge; target prediction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A comprehensive profile of the tilapia (Oreochromis niloticus) circular RNA and circRNA-miRNA network in the pathogenesis of meningoencephalitis of teleosts.Mol Omics. 2019 Jun 1;15(3):233-246. doi: 10.1039/c9mo00025a. Epub 2019 May 17. Mol Omics. 2019. PMID: 31098608
-
Identification of a circRNA-miRNA-mRNA network to explore the effects of circRNAs on pathogenesis and treatment of spinal cord injury.Life Sci. 2020 Sep 15;257:118039. doi: 10.1016/j.lfs.2020.118039. Epub 2020 Jul 2. Life Sci. 2020. PMID: 32621925
-
Unearthing Regulatory Axes of Breast Cancer circRNAs Networks to Find Novel Targets and Fathom Pivotal Mechanisms.Interdiscip Sci. 2019 Dec;11(4):711-722. doi: 10.1007/s12539-019-00339-6. Epub 2019 Jun 11. Interdiscip Sci. 2019. PMID: 31187432
-
Circular RNAs Act as miRNA Sponges.Adv Exp Med Biol. 2018;1087:67-79. doi: 10.1007/978-981-13-1426-1_6. Adv Exp Med Biol. 2018. PMID: 30259358 Review.
-
Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis.Genomics. 2020 Mar;112(2):1680-1685. doi: 10.1016/j.ygeno.2019.10.006. Epub 2019 Oct 15. Genomics. 2020. PMID: 31626900 Review.
Cited by
-
Circr, a Computational Tool to Identify miRNA:circRNA Associations.Front Bioinform. 2022 Mar 11;2:852834. doi: 10.3389/fbinf.2022.852834. eCollection 2022. Front Bioinform. 2022. PMID: 36304313 Free PMC article.
-
Astragalus IV Undermines Multi-Drug Resistance and Glycolysis of MDA-MB-231/ADR Cell Line by Depressing hsa_circ_0001982-miR-206/miR-613 Axis.Cancer Manag Res. 2021 Jul 22;13:5821-5833. doi: 10.2147/CMAR.S297008. eCollection 2021. Cancer Manag Res. 2021. PMID: 34326666 Free PMC article.
-
Circular RNA circGSK3B Promotes Cell Proliferation, Migration, and Invasion by Sponging miR-1265 and Regulating CAB39 Expression in Hepatocellular Carcinoma.Front Oncol. 2020 Nov 11;10:598256. doi: 10.3389/fonc.2020.598256. eCollection 2020. Front Oncol. 2020. PMID: 33262952 Free PMC article.
-
Identification of Specific Circular RNA Expression Patterns and MicroRNA Interaction Networks in Mesial Temporal Lobe Epilepsy.Front Genet. 2020 Sep 25;11:564301. doi: 10.3389/fgene.2020.564301. eCollection 2020. Front Genet. 2020. PMID: 33101384 Free PMC article.
-
Use of Circular RNAs in Diagnosis, Prognosis and Therapeutics of Renal Cell Carcinoma.Front Cell Dev Biol. 2022 Jun 22;10:879814. doi: 10.3389/fcell.2022.879814. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35813211 Free PMC article. Review.
References
-
- Hacker A., Capel B., Goodfellow P., Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

