Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model
- PMID: 31434879
- PMCID: PMC6704256
- DOI: 10.1038/s41467-019-11674-z
Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model
Abstract
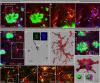
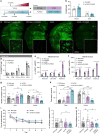
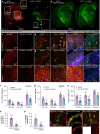
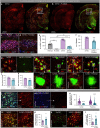
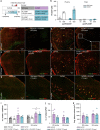
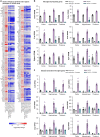
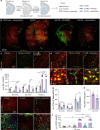
Many risk genes for the development of Alzheimer's disease (AD) are exclusively or highly expressed in myeloid cells. Microglia are dependent on colony-stimulating factor 1 receptor (CSF1R) signaling for their survival. We designed and synthesized a highly selective brain-penetrant CSF1R inhibitor (PLX5622) allowing for extended and specific microglial elimination, preceding and during pathology development. We find that in the 5xFAD mouse model of AD, plaques fail to form in the parenchymal space following microglial depletion, except in areas containing surviving microglia. Instead, Aβ deposits in cortical blood vessels reminiscent of cerebral amyloid angiopathy. Altered gene expression in the 5xFAD hippocampus is also reversed by the absence of microglia. Transcriptional analyses of the residual plaque-forming microglia show they exhibit a disease-associated microglia profile. Collectively, we describe the structure, formulation, and efficacy of PLX5622, which allows for sustained microglial depletion and identify roles of microglia in initiating plaque pathogenesis.
Conflict of interest statement
P.L.Severson, J.Z., E.A.B., Y.Z., W.S., J.L., G.H., A.R., G.T., J.W., M.N., P.Singh, S.B., P.I., C.Z., G.B., and B.L.W. are employees of Plexxikon Inc.; the remaining authors declare no competing interests.
Figures



Similar articles
-
Microglia depletion rapidly and reversibly alters amyloid pathology by modification of plaque compaction and morphologies.Neurobiol Dis. 2020 Aug;142:104956. doi: 10.1016/j.nbd.2020.104956. Epub 2020 May 30. Neurobiol Dis. 2020. PMID: 32479996 Free PMC article.
-
Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice.J Neuroinflammation. 2015 Aug 1;12:139. doi: 10.1186/s12974-015-0366-9. J Neuroinflammation. 2015. PMID: 26232154 Free PMC article.
-
Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer's disease.Mol Neurodegener. 2018 Mar 1;13(1):11. doi: 10.1186/s13024-018-0244-x. Mol Neurodegener. 2018. PMID: 29490706 Free PMC article.
-
Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer's-like pathology.Brain. 2016 Mar;139(Pt 3):891-907. doi: 10.1093/brain/awv379. Epub 2016 Jan 8. Brain. 2016. PMID: 26747862 Free PMC article.
-
Should We Open Fire on Microglia? Depletion Models as Tools to Elucidate Microglial Role in Health and Alzheimer's Disease.Int J Mol Sci. 2021 Sep 8;22(18):9734. doi: 10.3390/ijms22189734. Int J Mol Sci. 2021. PMID: 34575898 Free PMC article. Review.
Cited by
-
Targeting Microglia-Synapse Interactions in Alzheimer's Disease.Int J Mol Sci. 2021 Feb 26;22(5):2342. doi: 10.3390/ijms22052342. Int J Mol Sci. 2021. PMID: 33652870 Free PMC article. Review.
-
The Bidirectional Relationship Between Sleep and Inflammation Links Traumatic Brain Injury and Alzheimer's Disease.Front Neurosci. 2020 Aug 25;14:894. doi: 10.3389/fnins.2020.00894. eCollection 2020. Front Neurosci. 2020. PMID: 32982677 Free PMC article. Review.
-
Long-term microglia depletion impairs synapse elimination and auditory brainstem function.Sci Rep. 2022 Nov 2;12(1):18521. doi: 10.1038/s41598-022-23250-5. Sci Rep. 2022. PMID: 36323869 Free PMC article.
-
Delayed microglial depletion after spinal cord injury reduces chronic inflammation and neurodegeneration in the brain and improves neurological recovery in male mice.Theranostics. 2020 Sep 14;10(25):11376-11403. doi: 10.7150/thno.49199. eCollection 2020. Theranostics. 2020. PMID: 33052221 Free PMC article.
-
Microglia shield the murine brain from damage mediated by the cytokines IL-6 and IFN-α.Front Immunol. 2022 Oct 28;13:1036799. doi: 10.3389/fimmu.2022.1036799. eCollection 2022. Front Immunol. 2022. PMID: 36389783 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

