Human Cytomegalovirus US28 Ligand Binding Activity Is Required for Latency in CD34+ Hematopoietic Progenitor Cells and Humanized NSG Mice
- PMID: 31431555
- PMCID: PMC6703429
- DOI: 10.1128/mBio.01889-19
Human Cytomegalovirus US28 Ligand Binding Activity Is Required for Latency in CD34+ Hematopoietic Progenitor Cells and Humanized NSG Mice
Abstract
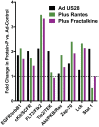
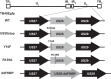
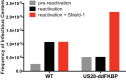
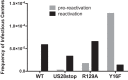
Human cytomegalovirus (HCMV) infection of CD34+ hematopoietic progenitor cells (CD34+ HPCs) provides a critical reservoir of virus in stem cell transplant patients, and viral reactivation remains a significant cause of morbidity and mortality. The HCMV chemokine receptor US28 is implicated in the regulation of viral latency and reactivation. To explore the role of US28 signaling in latency and reactivation, we analyzed protein tyrosine kinase signaling in CD34+ HPCs expressing US28. US28-ligand signaling in CD34+ HPCs induced changes in key regulators of cellular activation and differentiation. In vitro latency and reactivation assays utilizing CD34+ HPCs indicated that US28 was required for viral reactivation but not latency establishment or maintenance. Similarly, humanized NSG mice (huNSG) infected with TB40E-GFP-US28stop failed to reactivate upon treatment with granulocyte-colony-stimulating factor, but viral genome levels were maintained. Interestingly, HCMV-mediated changes in hematopoiesis during latency in vivo and in vitro was also dependent upon US28, as US28 directly promoted differentiation toward the myeloid lineage. To determine whether US28 constitutive activity and/or ligand-binding activity were required for latency and reactivation, we infected both huNSG mice and CD34+ HPCs in vitro with HCMV TB40E-GFP containing the US28-R129A mutation (no CA) or Y16F mutation (no ligand binding). TB40E-GFP-US28-R129A was maintained during latency and exhibited normal reactivation kinetics. In contrast, TB40E-GFP-US28-Y16F exhibited high levels of viral genome during latency and reactivation, indicating that the virus did not establish latency. These data indicate that US28 is necessary for viral reactivation and ligand binding activity is required for viral latency, highlighting the complex role of US28 during HCMV latency and reactivation.IMPORTANCE Human cytomegalovirus (HCMV) can establish latency following infection of CD34+ hematopoietic progenitor cells (HPCs), and reactivation from latency is a significant cause of viral disease and accelerated graft failure in bone marrow and solid-organ transplant patients. The precise molecular mechanisms of HCMV infection in HPCs are not well defined; however, select viral gene products are known to regulate aspects of latency and reactivation. The HCMV-encoded chemokine receptor US28, which binds multiple CC chemokines as well as CX3CR1, is expressed both during latent and lytic phases of the virus life cycle and plays a role in latency and reactivation. However, the specific timing of US28 expression and the role of ligand binding in these processes are not well defined. In this report, we determined that US28 is required for reactivation but not for maintaining latency. However, when present during latency, US28 ligand binding activity is critical to maintaining the virus in a quiescent state. We attribute the regulation of both latency and reactivation to the role of US28 in promoting myeloid lineage cell differentiation. These data highlight the dynamic and multifunctional nature of US28 during HCMV latency and reactivation.
Keywords: US28; hematopoiesis; human cytomegalovirus; latency; reactivation.
Copyright © 2019 Crawford et al.
Figures




Similar articles
-
Human Cytomegalovirus US28 Is Important for Latent Infection of Hematopoietic Progenitor Cells.J Virol. 2015 Dec 30;90(6):2959-70. doi: 10.1128/JVI.02507-15. J Virol. 2015. PMID: 26719258 Free PMC article.
-
Human Cytomegalovirus Requires Epidermal Growth Factor Receptor Signaling To Enter and Initiate the Early Steps in the Establishment of Latency in CD34+ Human Progenitor Cells.J Virol. 2017 Feb 14;91(5):e01206-16. doi: 10.1128/JVI.01206-16. Print 2017 Mar 1. J Virol. 2017. PMID: 27974567 Free PMC article.
-
CD34+ Hematopoietic Progenitor Cell Subsets Exhibit Differential Ability To Maintain Human Cytomegalovirus Latency and Persistence.J Virol. 2021 Jan 13;95(3):e02105-20. doi: 10.1128/JVI.02105-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177198 Free PMC article.
-
Advances in Model Systems for Human Cytomegalovirus Latency and Reactivation.mBio. 2022 Feb 22;13(1):e0172421. doi: 10.1128/mbio.01724-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012351 Free PMC article. Review.
-
HCMV latency: what regulates the regulators?Med Microbiol Immunol. 2019 Aug;208(3-4):431-438. doi: 10.1007/s00430-019-00581-1. Epub 2019 Feb 14. Med Microbiol Immunol. 2019. PMID: 30761409 Free PMC article. Review.
Cited by
-
Virus-Specific Nanobody-Chimeras Degrade the Human Cytomegalovirus US28 Protein in CD34+ Cells.Pathogens. 2024 Sep 24;13(10):821. doi: 10.3390/pathogens13100821. Pathogens. 2024. PMID: 39452693 Free PMC article.
-
Delivery of US28 by incoming HCMV particles rapidly attenuates Akt activity to suppress HCMV lytic replication in monocytes.Sci Signal. 2024 Aug 27;17(851):eadn8727. doi: 10.1126/scisignal.adn8727. Epub 2024 Aug 27. Sci Signal. 2024. PMID: 39190708 Free PMC article.
-
cGAS-STING-TBK1 Signaling Promotes Valproic Acid-Responsive Human Cytomegalovirus Immediate-Early Transcription during Infection of Incompletely Differentiated Myeloid Cells.Viruses. 2024 May 30;16(6):877. doi: 10.3390/v16060877. Viruses. 2024. PMID: 38932169 Free PMC article.
-
Proximity-dependent mapping of the HCMV US28 interactome identifies RhoGEF signaling as a requirement for efficient viral reactivation.PLoS Pathog. 2023 Oct 2;19(10):e1011682. doi: 10.1371/journal.ppat.1011682. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37782657 Free PMC article.
-
Mouse Models for Human Herpesviruses.Pathogens. 2023 Jul 19;12(7):953. doi: 10.3390/pathogens12070953. Pathogens. 2023. PMID: 37513800 Free PMC article. Review.
References
-
- Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. 2002. High risk of death due to bacterial and fungal infection among cytomegalovirus (CMV)-seronegative recipients of stem cell transplants from seropositive donors: evidence for indirect effects of primary CMV infection. J Infect Dis 185:273–282. doi:10.1086/338624. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
