The draft genome sequence of the spider Dysdera silvatica (Araneae, Dysderidae): A valuable resource for functional and evolutionary genomic studies in chelicerates
- PMID: 31430368
- PMCID: PMC6701490
- DOI: 10.1093/gigascience/giz099
The draft genome sequence of the spider Dysdera silvatica (Araneae, Dysderidae): A valuable resource for functional and evolutionary genomic studies in chelicerates
Abstract
Background: We present the draft genome sequence of Dysdera silvatica, a nocturnal ground-dwelling spider from a genus that has undergone a remarkable adaptive radiation in the Canary Islands.
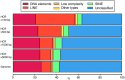
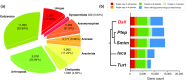
Results: The draft assembly was obtained using short (Illumina) and long (PaciBio and Nanopore) sequencing reads. Our de novo assembly (1.36 Gb), which represents 80% of the genome size estimated by flow cytometry (1.7 Gb), is constituted by a high fraction of interspersed repetitive elements (53.8%). The assembly completeness, using BUSCO and core eukaryotic genes, ranges from 90% to 96%. Functional annotations based on both ab initio and evidence-based information (including D. silvatica RNA sequencing) yielded a total of 48,619 protein-coding sequences, of which 36,398 (74.9%) have the molecular hallmark of known protein domains, or sequence similarity with Swiss-Prot sequences. The D. silvatica assembly is the first representative of the superfamily Dysderoidea, and just the second available genome of Synspermiata, one of the major evolutionary lineages of the "true spiders" (Araneomorphae).
Conclusions: Dysderoids, which are known for their numerous instances of adaptation to underground environments, include some of the few examples of trophic specialization within spiders and are excellent models for the study of cryptic female choice. This resource will be therefore useful as a starting point to study fundamental evolutionary and functional questions, including the molecular bases of the adaptation to extreme environments and ecological shifts, as well of the origin and evolution of relevant spider traits, such as the venom and silk.
Keywords: Araneomorphae; Canary Islands; genome annotation; hybrid genome assembly.
© The Author(s) 2019. Published by Oxford University Press.
Figures


Similar articles
-
The chromosome-scale assembly of the Canary Islands endemic spider Dysdera silvatica (Arachnida, Araneae) sheds light on the origin and genome structure of chemoreceptor gene families in chelicerates.Mol Ecol Resour. 2022 Jan;22(1):375-390. doi: 10.1111/1755-0998.13471. Epub 2021 Aug 5. Mol Ecol Resour. 2022. PMID: 34268885
-
Population Genomics of Adaptive Radiations: Exceptionally High Levels of Genetic Diversity and Recombination in an Endemic Spider From the Canary Islands.Mol Ecol. 2024 Nov;33(22):e17547. doi: 10.1111/mec.17547. Epub 2024 Oct 14. Mol Ecol. 2024. PMID: 39400446
-
A targeted gene phylogenetic framework to investigate diversification in the highly diverse yet geographically restricted red devil spiders (Araneae, Dysderidae).Cladistics. 2024 Dec;40(6):577-597. doi: 10.1111/cla.12595. Epub 2024 Aug 6. Cladistics. 2024. PMID: 39105704
-
Advances in the reconstruction of the spider tree of life: A roadmap for spider systematics and comparative studies.Cladistics. 2023 Dec;39(6):479-532. doi: 10.1111/cla.12557. Epub 2023 Oct 3. Cladistics. 2023. PMID: 37787157 Review.
-
Recent progress and prospects for advancing arachnid genomics.Curr Opin Insect Sci. 2018 Feb;25:51-57. doi: 10.1016/j.cois.2017.11.005. Epub 2017 Nov 21. Curr Opin Insect Sci. 2018. PMID: 29602362 Free PMC article. Review.
Cited by
-
Editorial.Dev Genes Evol. 2020 Mar;230(2):47-48. doi: 10.1007/s00427-020-00658-5. Dev Genes Evol. 2020. PMID: 32124013 No abstract available.
-
A chromosome-level genome of the spider Trichonephila antipodiana reveals the genetic basis of its polyphagy and evidence of an ancient whole-genome duplication event.Gigascience. 2021 Mar 19;10(3):giab016. doi: 10.1093/gigascience/giab016. Gigascience. 2021. PMID: 33739402 Free PMC article.
-
Genomic resources of aquatic Lepidoptera, Elophila obliteralis and Hyposmocoma kahamanoa, reveal similarities with Trichoptera in amino acid composition of major silk genes.G3 (Bethesda). 2024 Sep 4;14(9):jkae093. doi: 10.1093/g3journal/jkae093. G3 (Bethesda). 2024. PMID: 38722626 Free PMC article.
-
Annotated Draft Genomes of Two Caddisfly Species Plectrocnemia conspersa CURTIS and Hydropsyche tenuis NAVAS (Insecta: Trichoptera).Genome Biol Evol. 2019 Dec 1;11(12):3445-3451. doi: 10.1093/gbe/evz264. Genome Biol Evol. 2019. PMID: 31774498 Free PMC article.
-
Evolution of the Spider Homeobox Gene Repertoire by Tandem and Whole Genome Duplication.Mol Biol Evol. 2023 Dec 1;40(12):msad239. doi: 10.1093/molbev/msad239. Mol Biol Evol. 2023. PMID: 37935059 Free PMC article.
References
-
- World Spider Catalog (2018). 2018. http://wsc.nmbe.ch. Accessed on April 2019.
-
- Pekár S, Toft S. Trophic specialisation in a predatory group: the case of prey-specialised spiders (Araneae). Biol Rev. 2015;90(3):744–61. - PubMed
-
- Hopkin SP, Martin MH. Assimilation of zinc, cadmium, lead, copper, and iron by the spider Dysdera crocata, a predator of woodlice. Bull Environ Contam Toxicol. 1985;34:183–87. - PubMed
-
- Pekár S, Líznarová E, Řezác M. Suitability of woodlice prey for generalist and specialist spider predators: a comparative study. Ecol Entomol. 2016;41(2):123–30.
-
- Toft S, Macías-Hernández N. Metabolic adaptations for isopod specialization in three species of Dysdera spiders from the Canary Islands. Physiol Entomol. 2017;42(2):191–98.

