RNA degradomes reveal substrates and importance for dark and nitrogen stress responses of Arabidopsis XRN4
- PMID: 31428786
- PMCID: PMC6755094
- DOI: 10.1093/nar/gkz712
RNA degradomes reveal substrates and importance for dark and nitrogen stress responses of Arabidopsis XRN4
Abstract
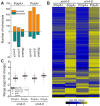
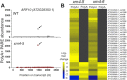
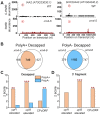
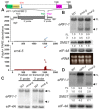
XRN4, the plant cytoplasmic homolog of yeast and metazoan XRN1, catalyzes exoribonucleolytic degradation of uncapped mRNAs from the 5' end. Most studies of cytoplasmic XRN substrates have focused on polyadenylated transcripts, although many substrates are likely first deadenylated. Here, we report the global investigation of XRN4 substrates in both polyadenylated and nonpolyadenylated RNA to better understand the impact of the enzyme in Arabidopsis. RNA degradome analysis demonstrated that xrn4 mutants overaccumulate many more decapped deadenylated intermediates than those that are polyadenylated. Among these XRN4 substrates that have 5' ends precisely at cap sites, those associated with photosynthesis, nitrogen responses and auxin responses were enriched. Moreover, xrn4 was found to be defective in the dark stress response and lateral root growth during N resupply, demonstrating that XRN4 is required during both processes. XRN4 also contributes to nonsense-mediated decay (NMD) and xrn4 accumulates 3' fragments of select NMD targets, despite the lack of the metazoan endoribonuclease SMG6 in plants. Beyond demonstrating that XRN4 is a major player in multiple decay pathways, this study identified intriguing molecular impacts of the enzyme, including those that led to new insights about mRNA decay and discovery of functional contributions at the whole-plant level.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Arabidopsis mRNA decay landscape shaped by XRN 5'-3' exoribonucleases.Plant J. 2023 May;114(4):895-913. doi: 10.1111/tpj.16181. Epub 2023 Mar 28. Plant J. 2023. PMID: 36987558
-
Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories.RNA. 2011 Mar;17(3):501-11. doi: 10.1261/rna.2467911. Epub 2011 Jan 11. RNA. 2011. PMID: 21224377 Free PMC article.
-
RNA degradome analysis reveals DNE1 endoribonuclease is required for the turnover of diverse mRNA substrates in Arabidopsis.Plant Cell. 2023 May 29;35(6):1936-1955. doi: 10.1093/plcell/koad085. Plant Cell. 2023. PMID: 37070465 Free PMC article.
-
XRN 5'→3' exoribonucleases: structure, mechanisms and functions.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):590-603. doi: 10.1016/j.bbagrm.2013.03.005. Epub 2013 Mar 19. Biochim Biophys Acta. 2013. PMID: 23517755 Free PMC article. Review.
-
NMD mechanism and the functions of Upf proteins in plant.Plant Cell Rep. 2016 Jan;35(1):5-15. doi: 10.1007/s00299-015-1867-9. Epub 2015 Sep 23. Plant Cell Rep. 2016. PMID: 26400685 Review.
Cited by
-
Monitoring mRNA Half-Life in Arabidopsis Using Droplet Digital PCR.Plants (Basel). 2022 Oct 5;11(19):2616. doi: 10.3390/plants11192616. Plants (Basel). 2022. PMID: 36235485 Free PMC article.
-
Nonsense-Mediated RNA Decay Factor UPF1 Is Critical for Posttranscriptional and Translational Gene Regulation in Arabidopsis.Plant Cell. 2020 Sep;32(9):2725-2741. doi: 10.1105/tpc.20.00244. Epub 2020 Jul 14. Plant Cell. 2020. PMID: 32665305 Free PMC article.
-
Comparative parallel analysis of RNA ends identifies mRNA substrates of a tRNA splicing endonuclease-initiated mRNA decay pathway.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2020429118. doi: 10.1073/pnas.2020429118. Proc Natl Acad Sci U S A. 2021. PMID: 33649230 Free PMC article.
-
SnRK2.10 kinase differentially modulates expression of hub WRKY transcription factors genes under salinity and oxidative stress in Arabidopsis thaliana.Front Plant Sci. 2023 Aug 9;14:1135240. doi: 10.3389/fpls.2023.1135240. eCollection 2023. Front Plant Sci. 2023. PMID: 37621885 Free PMC article.
-
Sugar Signaling and Post-transcriptional Regulation in Plants: An Overlooked or an Emerging Topic?Front Plant Sci. 2020 Nov 5;11:578096. doi: 10.3389/fpls.2020.578096. eCollection 2020. Front Plant Sci. 2020. PMID: 33224165 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

