The Fab portion of immunoglobulin G contributes to its binding to Fcγ receptor III
- PMID: 31420591
- PMCID: PMC6697678
- DOI: 10.1038/s41598-019-48323-w
The Fab portion of immunoglobulin G contributes to its binding to Fcγ receptor III
Abstract
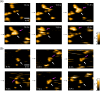
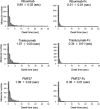
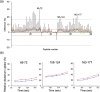
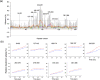
Most cells active in the immune system express receptors for antibodies which mediate a variety of defensive mechanisms. These receptors interact with the Fc portion of the antibody and are therefore collectively called Fc receptors. Here, using high-speed atomic force microscopy, we observe interactions of human, humanized, and mouse/human-chimeric immunoglobulin G1 (IgG1) antibodies and their cognate Fc receptor, FcγRIIIa. Our results demonstrate that not only Fc but also Fab positively contributes to the interaction with the receptor. Furthermore, hydrogen/deuterium exchange mass spectrometric analysis reveals that the Fab portion of IgG1 is directly involved in its interaction with FcγRIIIa, in addition to the canonical Fc-mediated interaction. By targeting the previously unidentified receptor-interaction sites in IgG-Fab, our findings could inspire therapeutic antibody engineering.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The Fab portion of immunoglobulin G has sites in the CL domain that interact with Fc gamma receptor IIIa.MAbs. 2022 Jan-Dec;14(1):2038531. doi: 10.1080/19420862.2022.2038531. MAbs. 2022. PMID: 35291930 Free PMC article.
-
Biophysical Characterization of the Contribution of the Fab Region to the IgG-FcγRIIIa Interaction.Biochemistry. 2023 Jan 17;62(2):262-269. doi: 10.1021/acs.biochem.1c00832. Epub 2022 May 23. Biochemistry. 2023. PMID: 35605982 Free PMC article.
-
Importance of the Side Chain at Position 296 of Antibody Fc in Interactions with FcγRIIIa and Other Fcγ Receptors.PLoS One. 2015 Oct 7;10(10):e0140120. doi: 10.1371/journal.pone.0140120. eCollection 2015. PLoS One. 2015. PMID: 26444434 Free PMC article.
-
Recognition of immunoglobulins by Fcgamma receptors.Mol Immunol. 2002 May;38(14):1073-83. doi: 10.1016/s0161-5890(02)00036-6. Mol Immunol. 2002. PMID: 11955599 Review.
-
Structural analysis of Fc/FcγR complexes: a blueprint for antibody design.Immunol Rev. 2015 Nov;268(1):201-21. doi: 10.1111/imr.12365. Immunol Rev. 2015. PMID: 26497522 Review.
Cited by
-
Factor VIII moiety of recombinant Factor VIII Fc fusion protein impacts Fc effector function and CD16+ NK cell activation.Front Immunol. 2024 Apr 9;15:1341013. doi: 10.3389/fimmu.2024.1341013. eCollection 2024. Front Immunol. 2024. PMID: 38655263 Free PMC article.
-
Quantitative Visualization of the Interaction between Complement Component C1 and Immunoglobulin G: The Effect of CH1 Domain Deletion.Int J Mol Sci. 2022 Feb 14;23(4):2090. doi: 10.3390/ijms23042090. Int J Mol Sci. 2022. PMID: 35216207 Free PMC article.
-
The Fab portion of immunoglobulin G has sites in the CL domain that interact with Fc gamma receptor IIIa.MAbs. 2022 Jan-Dec;14(1):2038531. doi: 10.1080/19420862.2022.2038531. MAbs. 2022. PMID: 35291930 Free PMC article.
-
Evaluation of strategies to modify Anti-SARS-CoV-2 monoclonal antibodies for optimal functionality as therapeutics.PLoS One. 2022 Jun 3;17(6):e0267796. doi: 10.1371/journal.pone.0267796. eCollection 2022. PLoS One. 2022. PMID: 35657812 Free PMC article.
-
Identification of Mispairing Omic Signatures in Chinese Hamster Ovary (CHO) Cells Producing a Tri-Specific Antibody.Biomedicines. 2023 Oct 25;11(11):2890. doi: 10.3390/biomedicines11112890. Biomedicines. 2023. PMID: 38001891 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

