Upper Respiratory Dysbiosis with a Facultative-dominated Ecotype in Advanced Lung Disease and Dynamic Change after Lung Transplant
- PMID: 31415219
- PMCID: PMC6945465
- DOI: 10.1513/AnnalsATS.201904-299OC
Upper Respiratory Dysbiosis with a Facultative-dominated Ecotype in Advanced Lung Disease and Dynamic Change after Lung Transplant
Abstract
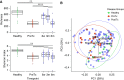
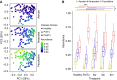
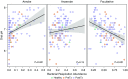
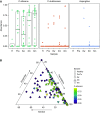
Rationale: The oropharyngeal microbiome is a primary source of lung microbiota, contributes to lower respiratory infection, and is also a driver of oral health.Objectives: We sought to understand oropharyngeal microbial communities in advanced lung disease, community dynamics after lung transplantation, and ecological features of dysbiosis.Methods: Oropharyngeal wash samples were obtained from individuals with end-stage disease awaiting transplantation (n = 22) and longitudinally from individuals at 6 weeks, 3 months, and 6 months after transplantation (n = 33), along with healthy control subjects (n = 14). Bacterial 16S and fungal internal transcribed spacer rRNA regions were deep-sequenced, and bacterial community respiratory patterns were imputed from taxonomic composition.Results: Healthy subjects' oropharyngeal microbiomes showed a gradient of community types reflecting relative enrichment of strictly anaerobic, aerobic, or facultative anaerobic bacteria. Patients with end-stage lung disease showed severe dysbiosis by both taxonomic composition and respiration phenotypes, with reduced richness and diversity, increased facultative and decreased aerobic bacteria, and absence of communities characterized by obligate aerobes. In patients at 6 weeks and 3 months post-transplant, richness and diversity were intermediate between healthy and pretransplant subjects, with near-normal distribution of community types. However, by 6 months post-transplant, oropharyngeal wash resembled the low-diversity facultative-dominated profile of pretransplant subjects. Community ecotype correlated with Candida abundance.Conclusions: End-stage lung disease is associated with marked upper respiratory tract dysbiosis involving both community structure and respiratory metabolism profiles of constituent bacteria. Dynamic changes occur after lung transplantation, with partial normalization early but later appearance of severe dysbiosis similar to pretransplant patients. Aberrant oropharyngeal communities may predispose to abnormal lung microbiota and infection risk both in advanced lung disease and after transplantation.
Keywords: bacteria; dysbiosis; fungi; lung transplantation; microbiome.
Figures

Similar articles
-
Signatures of COVID-19 Severity and Immune Response in the Respiratory Tract Microbiome.mBio. 2021 Aug 31;12(4):e0177721. doi: 10.1128/mBio.01777-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399607 Free PMC article.
-
The lung microbiome in lung transplantation.J Heart Lung Transplant. 2021 Aug;40(8):733-744. doi: 10.1016/j.healun.2021.04.014. Epub 2021 May 7. J Heart Lung Transplant. 2021. PMID: 34120840 Free PMC article. Review.
-
Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant.Am J Respir Crit Care Med. 2012 Sep 15;186(6):536-45. doi: 10.1164/rccm.201204-0693OC. Epub 2012 Jul 12. Am J Respir Crit Care Med. 2012. PMID: 22798321 Free PMC article.
-
Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients.ISME J. 2016 Jan;10(1):97-108. doi: 10.1038/ismej.2015.99. Epub 2015 Jul 7. ISME J. 2016. PMID: 26151645 Free PMC article.
-
Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery.Curr Opin Microbiol. 2018 Aug;44:34-40. doi: 10.1016/j.mib.2018.07.003. Epub 2018 Jul 20. Curr Opin Microbiol. 2018. PMID: 30036705 Free PMC article. Review.
Cited by
-
Signatures of COVID-19 Severity and Immune Response in the Respiratory Tract Microbiome.mBio. 2021 Aug 31;12(4):e0177721. doi: 10.1128/mBio.01777-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399607 Free PMC article.
-
Signatures of COVID-19 severity and immune response in the respiratory tract microbiome.medRxiv [Preprint]. 2021 Apr 5:2021.04.02.21254514. doi: 10.1101/2021.04.02.21254514. medRxiv. 2021. Update in: mBio. 2021 Aug 31;12(4):e0177721. doi: 10.1128/mBio.01777-21 PMID: 33851179 Free PMC article. Updated. Preprint.
-
The lung microbiome in lung transplantation.J Heart Lung Transplant. 2021 Aug;40(8):733-744. doi: 10.1016/j.healun.2021.04.014. Epub 2021 May 7. J Heart Lung Transplant. 2021. PMID: 34120840 Free PMC article. Review.
-
Unassigning bacterial species for microbiome studies.mSystems. 2024 Jul 23;9(7):e0051524. doi: 10.1128/msystems.00515-24. Epub 2024 Jun 24. mSystems. 2024. PMID: 38912768 Free PMC article.
-
The Lung Allograft Microbiome Associates with Pepsin, Inflammation, and Primary Graft Dysfunction.Am J Respir Crit Care Med. 2022 Dec 15;206(12):1508-1521. doi: 10.1164/rccm.202112-2786OC. Am J Respir Crit Care Med. 2022. PMID: 36103583 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

