Impact of the Position of the Chemically Modified 5-Furyl-2'-Deoxyuridine Nucleoside on the Thrombin DNA Aptamer-Protein Complex: Structural Insights into Aptamer Response from MD Simulations
- PMID: 31405145
- PMCID: PMC6720718
- DOI: 10.3390/molecules24162908
Impact of the Position of the Chemically Modified 5-Furyl-2'-Deoxyuridine Nucleoside on the Thrombin DNA Aptamer-Protein Complex: Structural Insights into Aptamer Response from MD Simulations
Abstract
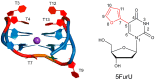
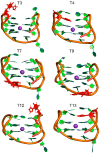
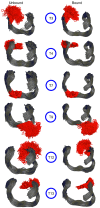
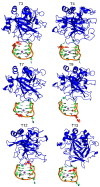
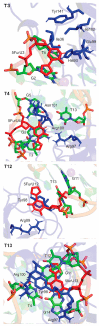
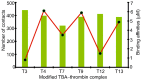
Aptamers are functional nucleic acids that bind to a range of targets (small molecules, proteins or cells) with a high affinity and specificity. Chemically-modified aptamers are of interest because the incorporation of novel nucleobase components can enhance aptamer binding to target proteins, while fluorescent base analogues permit the design of functional aptasensors that signal target binding. However, since optimally modified nucleoside designs have yet to be identified, information about how to fine tune aptamer stability and target binding affinity is required. The present work uses molecular dynamics (MD) simulations to investigate modifications to the prototypical thrombin-binding aptamer (TBA), which is a 15-mer DNA sequence that folds into a G-quadruplex structure connected by two TT loops and one TGT loop. Specifically, we modeled a previously synthesized thymine (T) analog, namely 5-furyl-2'-deoxyuridine (5FurU), into each of the six aptamer locations occupied by a thymine base in the TT or TGT loops of unbound and thrombin bound TBA. This modification and aptamer combination were chosen as a proof-of-principle because previous experimental studies have shown that TBA displays emissive sensitivity to target binding based on the local environment polarity at different 5FurU modification sites. Our simulations reveal that the chemically-modified base imparts noticeable structural changes to the aptamer without affecting the global conformation. Depending on the modification site, 5FurU performance is altered due to changes in the local environment, including the modification site structural dynamics, degree of solvent exposure, stacking with neighboring bases, and interactions with thrombin. Most importantly, these changes directly correlate with the experimentally-observed differences in the stability, binding affinity and emissive response of the modified aptamers. Therefore, the computational protocols implemented in the present work can be used in subsequent studies in a predictive way to aid the fine tuning of aptamer target recognition for use as biosensors (aptasensors) and/or therapeutics.
Keywords: G-quadruplexes; Thrombin–binding aptamer; chemical modification; computational modeling; fluorescent probes; protein binding..
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.Nucleic Acids Res. 2018 May 18;46(9):4819-4830. doi: 10.1093/nar/gky268. Nucleic Acids Res. 2018. PMID: 29684204 Free PMC article.
-
Manipulation of a DNA aptamer-protein binding site through arylation of internal guanine residues.Org Biomol Chem. 2018 May 23;16(20):3831-3840. doi: 10.1039/c8ob00704g. Org Biomol Chem. 2018. PMID: 29745412
-
Unlocking precision in aptamer engineering: a case study of the thrombin binding aptamer illustrates why modification size, quantity, and position matter.Nucleic Acids Res. 2024 Oct 14;52(18):10823-10835. doi: 10.1093/nar/gkae729. Nucleic Acids Res. 2024. PMID: 39217472 Free PMC article.
-
Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications.Curr Pharm Des. 2012;18(14):2036-47. doi: 10.2174/138161212799958387. Curr Pharm Des. 2012. PMID: 22376107 Review.
-
G-quadruplex-based aptamers targeting human thrombin: Discovery, chemical modifications and antithrombotic effects.Pharmacol Ther. 2021 Jan;217:107649. doi: 10.1016/j.pharmthera.2020.107649. Epub 2020 Aug 7. Pharmacol Ther. 2021. PMID: 32777331 Review.
Cited by
-
Aptamer-based Emerging Tools for Viral Biomarker Detection: A Focus on SARS-CoV-2.Curr Med Chem. 2023;30(8):910-934. doi: 10.2174/1568009622666220214101059. Curr Med Chem. 2023. PMID: 35156569 Review.
-
AptaNet as a deep learning approach for aptamer-protein interaction prediction.Sci Rep. 2021 Mar 16;11(1):6074. doi: 10.1038/s41598-021-85629-0. Sci Rep. 2021. PMID: 33727685 Free PMC article.
-
Discovery, Design, Synthesis, and Application of Nucleoside/Nucleotides.Molecules. 2020 Mar 27;25(7):1526. doi: 10.3390/molecules25071526. Molecules. 2020. PMID: 32230805 Free PMC article.
-
Exosite Binding in Thrombin: A Global Structural/Dynamic Overview of Complexes with Aptamers and Other Ligands.Int J Mol Sci. 2021 Oct 6;22(19):10803. doi: 10.3390/ijms221910803. Int J Mol Sci. 2021. PMID: 34639143 Free PMC article. Review.
-
Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication.Molecules. 2021 Feb 11;26(4):956. doi: 10.3390/molecules26040956. Molecules. 2021. PMID: 33670247 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

