Histone acetyltransferase GCN5-mediated regulation of long non-coding RNA At4 contributes to phosphate starvation response in Arabidopsis
- PMID: 31401648
- PMCID: PMC6859718
- DOI: 10.1093/jxb/erz359
Histone acetyltransferase GCN5-mediated regulation of long non-coding RNA At4 contributes to phosphate starvation response in Arabidopsis
Abstract
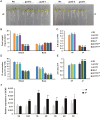
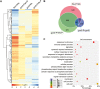
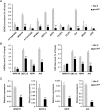
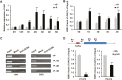
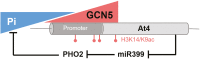
Phosphate availability is becoming a limiting environmental factor that inhibits plant growth and development. Here, we demonstrated that mutation of the histone acetyltransferase GCN5 impaired phosphate starvation responses (PSRs) in Arabidopsis. Transcriptome analysis revealed that 888 GCN5-regulated candidate genes were potentially involved in responding to phosphate starvation. ChIP assay indicated that four genes, including a long non-coding RNA (lncRNA) At4, are direct targets of GCN5 in PSR regulation. In addition, GCN5-mediated H3K9/14 acetylation of At4 determined dynamic At4 expression. Consistent with the function of At4 in phosphate distribution, mutation of GCN5 impaired phosphate accumulation between shoots and roots under phosphate deficiency condition, whereas constitutive expression of At4 in gcn5 mutants partially restored phosphate relocation. Further evidence proved that GCN5 regulation of At4 influenced the miRNA miR399 and its target PHO2 mRNA level. Taken together, we propose that GCN5-mediated histone acetylation plays a crucial role in PSR regulation via the At4-miR399-PHO2 pathway and provides a new epigenetic mechanism for the regulation of lncRNA in plants.
Keywords: Arabidopsis thaliana; At4; GCN5; histone acetylation; lncRNA; phosphate starvation response.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures
Similar articles
-
Updated Mechanisms of GCN5-The Monkey King of the Plant Kingdom in Plant Development and Resistance to Abiotic Stresses.Cells. 2021 Apr 22;10(5):979. doi: 10.3390/cells10050979. Cells. 2021. PMID: 33922251 Free PMC article. Review.
-
Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis.Plant J. 2015 Dec;84(6):1178-91. doi: 10.1111/tpj.13076. Plant J. 2015. PMID: 26576681
-
GENERAL CONTROL NONREPRESSED PROTEIN5-Mediated Histone Acetylation of FERRIC REDUCTASE DEFECTIVE3 Contributes to Iron Homeostasis in Arabidopsis.Plant Physiol. 2015 Aug;168(4):1309-20. doi: 10.1104/pp.15.00397. Epub 2015 May 22. Plant Physiol. 2015. PMID: 26002909 Free PMC article.
-
Synergistic action of histone acetyltransferase GCN5 and receptor CLAVATA1 negatively affects ethylene responses in Arabidopsis thaliana.J Exp Bot. 2016 Feb;67(3):905-18. doi: 10.1093/jxb/erv503. Epub 2015 Nov 23. J Exp Bot. 2016. PMID: 26596766
-
Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis.Mol Plant. 2010 Jul;3(4):670-7. doi: 10.1093/mp/ssq018. Epub 2010 May 9. Mol Plant. 2010. PMID: 20457643 Review.
Cited by
-
Updated Mechanisms of GCN5-The Monkey King of the Plant Kingdom in Plant Development and Resistance to Abiotic Stresses.Cells. 2021 Apr 22;10(5):979. doi: 10.3390/cells10050979. Cells. 2021. PMID: 33922251 Free PMC article. Review.
-
Epigenetic regulation: another layer in plant nutrition.Plant Signal Behav. 2020;15(1):1686236. doi: 10.1080/15592324.2019.1686236. Epub 2019 Nov 1. Plant Signal Behav. 2020. PMID: 31674259 Free PMC article. Review.
-
The Histone Acetyltransferase GCN5 and the Associated Coactivators ADA2: From Evolution of the SAGA Complex to the Biological Roles in Plants.Plants (Basel). 2021 Feb 5;10(2):308. doi: 10.3390/plants10020308. Plants (Basel). 2021. PMID: 33562796 Free PMC article. Review.
-
Genome-wide analysis of long non-coding RNAs (lncRNAs) in two contrasting soybean genotypes subjected to phosphate starvation.BMC Genomics. 2021 Jun 9;22(1):433. doi: 10.1186/s12864-021-07750-8. BMC Genomics. 2021. PMID: 34107875 Free PMC article.
-
A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity.Nat Commun. 2020 Feb 13;11(1):870. doi: 10.1038/s41467-020-14558-9. Nat Commun. 2020. PMID: 32054839 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

