Identification of the Capsid Binding Site in the Herpes Simplex Virus 1 Nuclear Egress Complex and Its Role in Viral Primary Envelopment and Replication
- PMID: 31391274
- PMCID: PMC6803286
- DOI: 10.1128/JVI.01290-19
Identification of the Capsid Binding Site in the Herpes Simplex Virus 1 Nuclear Egress Complex and Its Role in Viral Primary Envelopment and Replication
Abstract
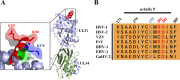
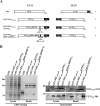
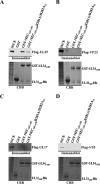
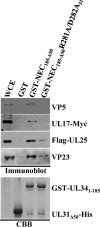
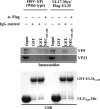
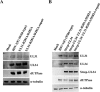
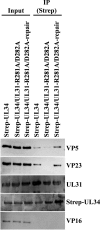
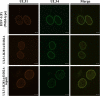
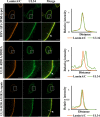
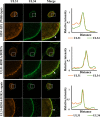
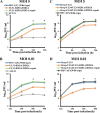
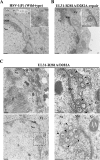
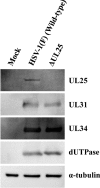
During nuclear egress of nascent progeny herpesvirus nucleocapsids, the nucleocapsids acquire a primary envelope by budding through the inner nuclear membrane of infected cells into the perinuclear space between the inner and outer nuclear membranes. Herpes simplex virus 1 (HSV-1) UL34 and UL31 proteins form a nuclear egress complex (NEC) and play critical roles in this budding process, designated primary envelopment. To clarify the role of NEC binding to progeny nucleocapsids in HSV-1 primary envelopment, we established an assay system for HSV-1 NEC binding to nucleocapsids and capsid proteins in vitro Using this assay system, we showed that HSV-1 NEC bound to nucleocapsids and to capsid protein UL25 but not to the other capsid proteins tested (i.e., VP5, VP23, and UL17) and that HSV-1 NEC binding of nucleocapsids was mediated by the interaction of NEC with UL25. UL31 residues arginine-281 (R281) and aspartic acid-282 (D282) were required for efficient NEC binding to nucleocapsids and UL25. We also showed that alanine substitution of UL31 R281 and D282 reduced HSV-1 replication, caused aberrant accumulation of capsids in the nucleus, and induced an accumulation of empty vesicles that were similar in size and morphology to primary envelopes in the perinuclear space. These results suggested that NEC binding via UL31 R281 and D282 to nucleocapsids, and probably to UL25 in the nucleocapsids, has an important role in HSV-1 replication by promoting the incorporation of nucleocapsids into vesicles during primary envelopment.IMPORTANCE Binding of HSV-1 NEC to nucleocapsids has been thought to promote nucleocapsid budding at the inner nuclear membrane and subsequent incorporation of nucleocapsids into vesicles during nuclear egress of nucleocapsids. However, data to directly support this hypothesis have not been reported thus far. In this study, we have present data showing that two amino acids in the membrane-distal face of the HSV-1 NEC, which contains the putative capsid binding site based on the solved NEC structure, were in fact required for efficient NEC binding to nucleocapsids and for efficient incorporation of nucleocapsids into vesicles during primary envelopment. This is the first report showing direct linkage between NEC binding to nucleocapsids and an increase in nucleocapsid incorporation into vesicles during herpesvirus primary envelopment.
Keywords: capsid; herpes simplex virus; nuclear egress.
Copyright © 2019 American Society for Microbiology.
Figures




Similar articles
-
Roles of the Interhexamer Contact Site for Hexagonal Lattice Formation of the Herpes Simplex Virus 1 Nuclear Egress Complex in Viral Primary Envelopment and Replication.J Virol. 2019 Jun 28;93(14):e00498-19. doi: 10.1128/JVI.00498-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31043535 Free PMC article.
-
Role of Host Cell p32 in Herpes Simplex Virus 1 De-Envelopment during Viral Nuclear Egress.J Virol. 2015 Sep;89(17):8982-98. doi: 10.1128/JVI.01220-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085152 Free PMC article.
-
Role of the Arginine Cluster in the Disordered Domain of Herpes Simplex Virus 1 UL34 for the Recruitment of ESCRT-III for Viral Primary Envelopment.J Virol. 2022 Jan 26;96(2):e0170421. doi: 10.1128/JVI.01704-21. Epub 2021 Nov 3. J Virol. 2022. PMID: 34730397 Free PMC article.
-
Host and Viral Factors Involved in Nuclear Egress of Herpes Simplex Virus 1.Viruses. 2021 Apr 25;13(5):754. doi: 10.3390/v13050754. Viruses. 2021. PMID: 33923040 Free PMC article. Review.
-
Virus Assembly and Egress of HSV.Adv Exp Med Biol. 2018;1045:23-44. doi: 10.1007/978-981-10-7230-7_2. Adv Exp Med Biol. 2018. PMID: 29896661 Review.
Cited by
-
Cell Culture Evolution of a Herpes Simplex Virus 1 (HSV-1)/Varicella-Zoster Virus (VZV) UL34/ORF24 Chimeric Virus Reveals Novel Functions for HSV Genes in Capsid Nuclear Egress.J Virol. 2021 Nov 9;95(23):e0095721. doi: 10.1128/JVI.00957-21. Epub 2021 Sep 15. J Virol. 2021. PMID: 34523964 Free PMC article.
-
Redundant and Specific Roles of A-Type Lamins and Lamin B Receptor in Herpes Simplex Virus 1 Infection.J Virol. 2022 Dec 21;96(24):e0142922. doi: 10.1128/jvi.01429-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448808 Free PMC article.
-
The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.Sci Adv. 2024 Feb 23;10(8):eadj1640. doi: 10.1126/sciadv.adj1640. Epub 2024 Feb 23. Sci Adv. 2024. PMID: 38394211 Free PMC article.
-
Structural basis for capsid recruitment and coat formation during HSV-1 nuclear egress.Elife. 2020 Jun 24;9:e56627. doi: 10.7554/eLife.56627. Elife. 2020. PMID: 32579107 Free PMC article.
-
Role of the Orphan Transporter SLC35E1 in the Nuclear Egress of Herpes Simplex Virus 1.J Virol. 2022 May 25;96(10):e0030622. doi: 10.1128/jvi.00306-22. Epub 2022 Apr 27. J Virol. 2022. PMID: 35475666 Free PMC article.
References
-
- Pellett PE, Roizman B. 2013. Herpesviridae, p 1802–1822. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott-Williams &Wilkins, Philadelphia, PA.
-
- Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott-Williams &Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

