Successful Invasions of Short Internally Deleted Elements (SIDEs) and Its Partner CR1 in Lepidoptera Insects
- PMID: 31384954
- PMCID: PMC6740152
- DOI: 10.1093/gbe/evz174
Successful Invasions of Short Internally Deleted Elements (SIDEs) and Its Partner CR1 in Lepidoptera Insects
Abstract
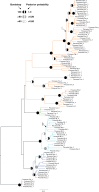
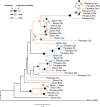
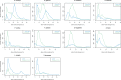
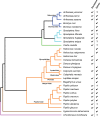
Although DNA transposons often generated internal deleted derivatives such as miniature inverted-repeat transposable elements, short internally deleted elements (SIDEs) derived from nonlong terminal-repeat retrotransposons are rare. Here, we found a novel SIDE, named Persaeus, that originated from the chicken repeat 1 (CR1) retrotransposon Zenon and it has been found widespread in Lepidoptera insects. Our findings suggested that Persaeus and the partner Zenon have experienced a transposition burst in their host genomes and the copy number of Persaeus and Zenon in assayed genomes are significantly correlated. Accordingly, the activity though age analysis indicated that the replication wave of Persaeus coincided with that of Zenon. Phylogenetic analyses suggested that Persaeus may have evolved at least four times independently, and that it has been vertically transferred into its host genomes. Together, our results provide new insights into the evolution dynamics of SIDEs and its partner non-LTRs.
Keywords: Lepidoptera; chicken repeat 1 (CR1); long interspersed element (LINE); short internally deleted element (SIDE); transposable elements evolutionary dynamics; vertical inheritance.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Vertical evolution and horizontal transfer of CR1 non-LTR retrotransposons and Tc1/mariner DNA transposons in Lepidoptera species.Mol Biol Evol. 2012 Dec;29(12):3685-702. doi: 10.1093/molbev/mss181. Epub 2012 Jul 23. Mol Biol Evol. 2012. PMID: 22826456
-
Differential Conservation and Loss of Chicken Repeat 1 (CR1) Retrotransposons in Squamates Reveal Lineage-Specific Genome Dynamics Across Reptiles.Genome Biol Evol. 2024 Aug 5;16(8):evae157. doi: 10.1093/gbe/evae157. Genome Biol Evol. 2024. PMID: 39031594 Free PMC article.
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
-
A retrotransposon of the non-long terminal repeat class from the human blood fluke Schistosoma mansoni. Similarities to the chicken-repeat-1-like elements of vertebrates.Mol Biol Evol. 1997 Jun;14(6):602-10. doi: 10.1093/oxfordjournals.molbev.a025799. Mol Biol Evol. 1997. PMID: 9190061
-
Evolutionary forces generating sequence homogeneity and heterogeneity within retrotransposon families.Cytogenet Genome Res. 2005;110(1-4):383-91. doi: 10.1159/000084970. Cytogenet Genome Res. 2005. PMID: 16093690 Review.
Cited by
-
Widespread HCD-tRNA derived SINEs in bivalves rely on multiple LINE partners and accumulate in genic regions.Mob DNA. 2024 Oct 16;15(1):22. doi: 10.1186/s13100-024-00332-x. Mob DNA. 2024. PMID: 39415259 Free PMC article.
References
-
- Bailly-Bechet M, Haudry A, Lerat E.. 2014. One code to find them all: a Perl tool to conveniently parse RepeatMasker output files. Mob DNA 5(1):13.
-
- Baker AJ, Haddrath O, McPherson JD, Cloutier A.. 2014. Genomic support for a moa-tinamou clade and adaptive morphological convergence in flightless ratites. Mol Biol Evol. 31(7):1686–1696. - PubMed
-
- Bibillo A, Eickbush T.. 2004. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 279(15):14945–14953. - PubMed
-
- Biedler J, Tu Z.. 2003. Non-LTR retrotransposons in the African malaria mosquito, Anopheles gambiae: unprecedented diversity and evidence of recent activity. Mol Biol Evol. 20(11):1811–1825. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

