Survival Analysis of Multi-Omics Data Identifies Potential Prognostic Markers of Pancreatic Ductal Adenocarcinoma
- PMID: 31379917
- PMCID: PMC6659773
- DOI: 10.3389/fgene.2019.00624
Survival Analysis of Multi-Omics Data Identifies Potential Prognostic Markers of Pancreatic Ductal Adenocarcinoma
Abstract
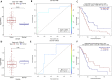
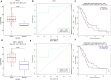
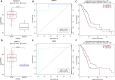
Pancreatic ductal adenocarcinoma (PDAC) is the most common and among the deadliest of pancreatic cancers. Its 5-year survival is only ∼8%. Pancreatic cancers are a heterogeneous group of diseases, of which PDAC is particularly aggressive. Like many other cancers, PDAC also starts as a pre-invasive precursor lesion (known as pancreatic intraepithelial neoplasia, PanIN), which offers an opportunity for both early detection and early treatment. Even advanced PDAC can benefit from prognostic biomarkers. However, reliable biomarkers for early diagnosis or those for prognosis of therapy remain an unfulfilled goal for PDAC. In this study, we selected 153 PDAC patients from the TCGA database and used their clinical, DNA methylation, gene expression, and micro-RNA (miRNA) and long non-coding RNA (lncRNA) expression data for multi-omics analysis. Differential methylations at about 12,000 CpG sites were observed in PDAC tumor genomes, with about 61% of them hypermethylated, predominantly in the promoter regions and in CpG-islands. We correlated promoter methylation and gene expression for mRNAs and identified 17 genes that were previously recognized as PDAC biomarkers. Similarly, several genes (B3GNT3, DMBT1, DEPDC1B) and lncRNAs (PVT1, and GATA6-AS) are strongly correlated with survival, which have not been reported in PDAC before. Other genes such as EFR3B, whose biological roles are not well known in mammals are also found to strongly associated with survival. We further identified 406 promoter methylation target loci associated with patients survival, including known esophageal squamous cell carcinoma biomarkers, cg03234186 (ZNF154), and cg02587316, cg18630667, and cg05020604 (ZNF382). Overall, this is one of the first studies that identified survival associated genes using multi-omics data from PDAC patients.
Keywords: DEG: differentially expressed gene; DMR: differentially methylated region; Dm-CpG: Differentially methylated CpG; FDR: false discovery rate; GDC: The Genomic Data Commons; HR: hazard ratio; TCGA: The Cancer Genome Atlas.
Figures

Similar articles
-
High methylation levels of PCDH10 predict poor prognosis in patients with pancreatic ductal adenocarcinoma.BMC Cancer. 2019 May 14;19(1):452. doi: 10.1186/s12885-019-5616-2. BMC Cancer. 2019. PMID: 31088413 Free PMC article.
-
DNA methylome in pancreatic cancer identified novel promoter hyper-methylation in NPY and FAIM2 genes associated with poor prognosis in Indian patient cohort.Cancer Cell Int. 2022 Nov 3;22(1):334. doi: 10.1186/s12935-022-02737-1. Cancer Cell Int. 2022. PMID: 36329447 Free PMC article.
-
Multi-omics analysis of intra-tumoural and inter-tumoural heterogeneity in pancreatic ductal adenocarcinoma.Clin Transl Med. 2022 Jan;12(1):e670. doi: 10.1002/ctm2.670. Clin Transl Med. 2022. PMID: 35061935 Free PMC article.
-
MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions.World J Gastrointest Oncol. 2016 Jan 15;8(1):18-29. doi: 10.4251/wjgo.v8.i1.18. World J Gastrointest Oncol. 2016. PMID: 26798434 Free PMC article. Review.
-
Molecular Biomarkers of Pancreatic Intraepithelial Neoplasia and Their Implications in Early Diagnosis and Therapeutic Intervention of Pancreatic Cancer.Int J Biol Sci. 2016 Jan 28;12(3):292-301. doi: 10.7150/ijbs.14995. eCollection 2016. Int J Biol Sci. 2016. PMID: 26929736 Free PMC article. Review.
Cited by
-
Identifying Drug Targets in Pancreatic Ductal Adenocarcinoma Through Machine Learning, Analyzing Biomolecular Networks, and Structural Modeling.Front Pharmacol. 2020 Apr 30;11:534. doi: 10.3389/fphar.2020.00534. eCollection 2020. Front Pharmacol. 2020. PMID: 32425783 Free PMC article.
-
Identification of Pan-Cancer Prognostic Biomarkers Through Integration of Multi-Omics Data.Front Bioeng Biotechnol. 2020 Apr 2;8:268. doi: 10.3389/fbioe.2020.00268. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32300588 Free PMC article.
-
Prognostic Nomogram for Patients With Pancreatic Ductal Adenocarcinoma of Pancreatic Head After Pancreaticoduodenectomy.Clin Med Insights Oncol. 2021 Jun 18;15:11795549211024149. doi: 10.1177/11795549211024149. eCollection 2021. Clin Med Insights Oncol. 2021. PMID: 34211308 Free PMC article.
-
Epigenetic Alterations in Pancreatic Cancer Metastasis.Biomolecules. 2021 Jul 22;11(8):1082. doi: 10.3390/biom11081082. Biomolecules. 2021. PMID: 34439749 Free PMC article. Review.
-
Genetics, Genomics and Emerging Molecular Therapies of Pancreatic Cancer.Cancers (Basel). 2023 Jan 27;15(3):779. doi: 10.3390/cancers15030779. Cancers (Basel). 2023. PMID: 36765737 Free PMC article. Review.
References
-
- Akbani R., Xhang N., Broom B. M. (2010). TCGA batch effect. [http://bioinformatics.mdanderson.org/tcgambatch/].
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources
Research Materials

