Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5
- PMID: 31379819
- PMCID: PMC6652118
- DOI: 10.3389/fimmu.2019.01586
Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5
Abstract
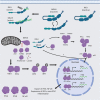
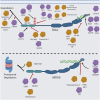
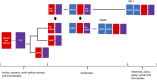
RIG-I (Retinoic acid-inducible gene I) and MDA5 (Melanoma Differentiation-Associated protein 5), collectively known as the RIG-I-like receptors (RLRs), are key protein sensors of the pathogen-associated molecular patterns (PAMPs) in the form of viral double-stranded RNA (dsRNA) motifs to induce expression of type 1 interferons (IFN1) (IFNα and IFNβ) and other pro-inflammatory cytokines during the early stage of viral infection. While RIG-I and MDA5 share many genetic, structural and functional similarities, there is increasing evidence that they can have significantly different strategies to recognize different pathogens, PAMPs, and in different host species. This review article discusses the similarities and differences between RIG-I and MDA5 from multiple perspectives, including their structures, evolution and functional relationships with other cellular proteins, their differential mechanisms of distinguishing between host and viral dsRNAs and interactions with host and viral protein factors, and their immunogenic signaling. A comprehensive comparative analysis can help inform future studies of RIG-I and MDA5 in order to fully understand their functions in order to optimize potential therapeutic approaches targeting them.
Keywords: CARD; MDA5; PAMP; PRRs; RIG-I; antiviral; inflammation; interferon.
Figures




Similar articles
-
An arms race between 5'ppp-RNA virus and its alternative recognition receptor MDA5 in RIG-I-lost teleost fish.Elife. 2024 Sep 30;13:RP94898. doi: 10.7554/eLife.94898. Elife. 2024. PMID: 39347580 Free PMC article.
-
Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses.Nature. 2006 May 4;441(7089):101-5. doi: 10.1038/nature04734. Epub 2006 Apr 9. Nature. 2006. PMID: 16625202
-
Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5.J Exp Med. 2008 Jul 7;205(7):1601-10. doi: 10.1084/jem.20080091. J Exp Med. 2008. PMID: 18591409 Free PMC article.
-
The anticancer functions of RIG-I-like receptors, RIG-I and MDA5, and their applications in cancer therapy.Transl Res. 2017 Dec;190:51-60. doi: 10.1016/j.trsl.2017.08.004. Epub 2017 Aug 31. Transl Res. 2017. PMID: 28917654 Review.
-
Immunogenicity of In Vitro-Transcribed RNA.Acc Chem Res. 2021 Nov 2;54(21):4012-4023. doi: 10.1021/acs.accounts.1c00521. Epub 2021 Oct 22. Acc Chem Res. 2021. PMID: 34677064 Free PMC article. Review.
Cited by
-
mRNA therapeutics: New vaccination and beyond.Fundam Res. 2023 Mar 16;3(5):749-759. doi: 10.1016/j.fmre.2023.02.022. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933291 Free PMC article. Review.
-
Porcine promyelocytic leukemia protein isoforms suppress Japanese encephalitis virus replication in PK15 cells.Virol J. 2023 Nov 29;20(1):280. doi: 10.1186/s12985-023-02212-x. Virol J. 2023. PMID: 38031162 Free PMC article.
-
Factors affecting RIG-I-Like receptors activation - New research direction for viral hemorrhagic fevers.Front Immunol. 2022 Sep 29;13:1010635. doi: 10.3389/fimmu.2022.1010635. eCollection 2022. Front Immunol. 2022. PMID: 36248895 Free PMC article. Review.
-
A Role for the Chicken Interferon-Stimulated Gene CMPK2 in the Host Response Against Virus Infection.Front Microbiol. 2022 May 11;13:874331. doi: 10.3389/fmicb.2022.874331. eCollection 2022. Front Microbiol. 2022. PMID: 35633731 Free PMC article.
-
Type I Interferon: Monkeypox/Mpox Viruses Achilles Heel?Adv Exp Med Biol. 2024;1451:125-137. doi: 10.1007/978-3-031-57165-7_8. Adv Exp Med Biol. 2024. PMID: 38801575 Review.
References
-
- Sun YW. RIG-I, a human homolog gene of RNA helicase, is induced by retinoic acid during the differentiation of acute promyelocytic leukemia cell. Biochem Biophys Res Commun. (1997) 292:274–9.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

