DNA mismatch repair is required for the host innate response and controls cellular fate after influenza virus infection
- PMID: 31358986
- PMCID: PMC6814535
- DOI: 10.1038/s41564-019-0509-3
DNA mismatch repair is required for the host innate response and controls cellular fate after influenza virus infection
Abstract
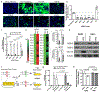
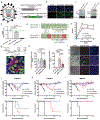
Despite the cytopathic nature of influenza A virus (IAV) replication, we recently reported that a subset of lung epithelial club cells is able to intrinsically clear the virus and survive infection. However, the mechanisms that drive cell survival during a normally lytic infection remained unclear. Using a loss-of-function screening approach, we discovered that the DNA mismatch repair (MMR) pathway is essential for club cell survival of IAV infection. Repair of virally induced oxidative damage by the DNA MMR pathway not only allowed cell survival of infection, but also facilitated host gene transcription, including the expression of antiviral and stress response genes. Enhanced viral suppression of the DNA MMR pathway prevented club cell survival and increased the severity of viral disease in vivo. Altogether, these results identify previously unappreciated roles for DNA MMR as a central modulator of cellular fate and a contributor to the innate antiviral response, which together control influenza viral disease severity.
Conflict of interest statement
COMPETING INTERESTS
Duke University has filed a provisional patent for targeting DNA MMR as a method to enhance the growth of influenza vaccine strains.
Figures




Similar articles
-
Cell type- and replication stage-specific influenza virus responses in vivo.PLoS Pathog. 2020 Aug 13;16(8):e1008760. doi: 10.1371/journal.ppat.1008760. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32790753 Free PMC article.
-
HDAC6 Restricts Influenza A Virus by Deacetylation of the RNA Polymerase PA Subunit.J Virol. 2019 Feb 5;93(4):e01896-18. doi: 10.1128/JVI.01896-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30518648 Free PMC article.
-
Identification of lncRNA-155 encoded by MIR155HG as a novel regulator of innate immunity against influenza A virus infection.Cell Microbiol. 2019 Aug;21(8):e13036. doi: 10.1111/cmi.13036. Epub 2019 May 29. Cell Microbiol. 2019. PMID: 31045320
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Role of microRNA and Oxidative Stress in Influenza A Virus Pathogenesis.Int J Mol Sci. 2020 Nov 25;21(23):8962. doi: 10.3390/ijms21238962. Int J Mol Sci. 2020. PMID: 33255826 Free PMC article. Review.
Cited by
-
Viral Modulation of the DNA Damage Response and Innate Immunity: Two Sides of the Same Coin.J Mol Biol. 2022 Mar 30;434(6):167327. doi: 10.1016/j.jmb.2021.167327. Epub 2021 Oct 22. J Mol Biol. 2022. PMID: 34695379 Free PMC article. Review.
-
Cilia-related gene signature in the nasal mucosa correlates with disease severity and outcomes in critical respiratory syncytial virus bronchiolitis.Front Immunol. 2022 Sep 23;13:924792. doi: 10.3389/fimmu.2022.924792. eCollection 2022. Front Immunol. 2022. PMID: 36211387 Free PMC article.
-
Delineating the SARS-CoV-2 Induced Interplay between the Host Immune System and the DNA Damage Response Network.Vaccines (Basel). 2022 Oct 20;10(10):1764. doi: 10.3390/vaccines10101764. Vaccines (Basel). 2022. PMID: 36298629 Free PMC article. Review.
-
Statistical Analysis of Common Respiratory Viruses Reveals the Binary of Virus-Virus Interaction.Microbiol Spectr. 2023 Aug 17;11(4):e0001923. doi: 10.1128/spectrum.00019-23. Epub 2023 Jun 28. Microbiol Spectr. 2023. PMID: 37378522 Free PMC article.
-
Deficient DNA mismatch repair and persistence of SARS-CoV-2 RNA shedding: a case report of hereditary nonpolyposis colorectal cancer with COVID-19 infection.BMC Infect Dis. 2021 Aug 21;21(1):854. doi: 10.1186/s12879-021-06500-1. BMC Infect Dis. 2021. PMID: 34418963 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

