Structural Basis for Target-Directed MicroRNA Degradation
- PMID: 31353209
- PMCID: PMC6754277
- DOI: 10.1016/j.molcel.2019.06.019
Structural Basis for Target-Directed MicroRNA Degradation
Abstract
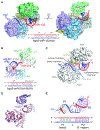
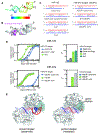
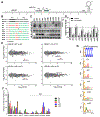
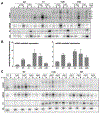
MicroRNAs (miRNAs) broadly regulate gene expression through association with Argonaute (Ago), which also protects miRNAs from degradation. However, miRNA stability is known to vary and is regulated by poorly understood mechanisms. A major emerging process, termed target-directed miRNA degradation (TDMD), employs specialized target RNAs to selectively bind to miRNAs and induce their decay. Here, we report structures of human Ago2 (hAgo2) bound to miRNAs and TDMD-inducing targets. miRNA and target form a bipartite duplex with an unpaired flexible linker. hAgo2 cannot physically accommodate the RNA, causing the duplex to bend at the linker and display the miRNA 3' end for enzymatic attack. Altering 3' end display by changing linker flexibility, changing 3' end complementarity, or mutationally inducing 3' end release impacts TDMD efficiency, leading to production of distinct 3'-miRNA isoforms in cells. Our results uncover the mechanism driving TDMD and reveal 3' end display as a key determinant regulating miRNA activity via 3' remodeling and/or degradation.
Keywords: Argonaute; HSUR1; TDMD; miRNA; miRNA-degradation; microRNA; tailing; trimming.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
Authors declare no competing interests.
Figures


Similar articles
-
MicroRNA turnover: a tale of tailing, trimming, and targets.Trends Biochem Sci. 2023 Jan;48(1):26-39. doi: 10.1016/j.tibs.2022.06.005. Epub 2022 Jul 7. Trends Biochem Sci. 2023. PMID: 35811249 Free PMC article. Review.
-
A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming.Science. 2020 Dec 18;370(6523):eabc9546. doi: 10.1126/science.abc9546. Epub 2020 Nov 12. Science. 2020. PMID: 33184234 Free PMC article.
-
Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins.Genes Dev. 2021 Dec 1;35(23-24):1595-1609. doi: 10.1101/gad.348874.121. Epub 2021 Nov 24. Genes Dev. 2021. PMID: 34819352 Free PMC article.
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
-
SnapShot: Target-directed miRNA degradation.Cell. 2023 Dec 7;186(25):5674-5674.e1. doi: 10.1016/j.cell.2023.11.020. Cell. 2023. PMID: 38065084
Cited by
-
LncRNA MIR200CHG inhibits EMT in gastric cancer by stabilizing miR-200c from target-directed miRNA degradation.Nat Commun. 2023 Dec 8;14(1):8141. doi: 10.1038/s41467-023-43974-w. Nat Commun. 2023. PMID: 38065939 Free PMC article.
-
What goes up must come down: off switches for regulatory RNAs.Genes Dev. 2024 Aug 20;38(13-14):597-613. doi: 10.1101/gad.351934.124. Genes Dev. 2024. PMID: 39111824 Free PMC article. Review.
-
MicroRNAs as monitoring markers for right-sided heart failure and congestive hepatopathy.J Med Life. 2021 Mar-Apr;14(2):142-147. doi: 10.25122/jml-2021-0071. J Med Life. 2021. PMID: 34104236 Free PMC article. Review.
-
The linear ANRIL transcript P14AS regulates the NF-κB signaling to promote colon cancer progression.Mol Med. 2023 Dec 1;29(1):162. doi: 10.1186/s10020-023-00761-z. Mol Med. 2023. PMID: 38041015 Free PMC article.
-
How Complementary Targets Expose the microRNA 3' End for Tailing and Trimming during Target-Directed microRNA Degradation.Cold Spring Harb Symp Quant Biol. 2019;84:179-183. doi: 10.1101/sqb.2019.84.039321. Epub 2020 Feb 4. Cold Spring Harb Symp Quant Biol. 2019. PMID: 32019864 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

