Synergistic inhibition of csal1 and csal3 in granulosa cell proliferation and steroidogenesis of hen ovarian prehierarchical development†
- PMID: 31350846
- PMCID: PMC6877779
- DOI: 10.1093/biolre/ioz137
Synergistic inhibition of csal1 and csal3 in granulosa cell proliferation and steroidogenesis of hen ovarian prehierarchical development†
Abstract
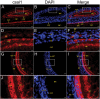
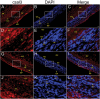
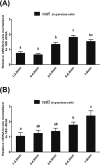
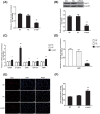
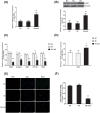
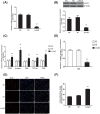
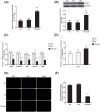
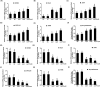
SALL1 and SALL3 are transcription factors that play an essential role in regulating developmental processes and organogenesis in many species. However, the functional role of SALL1 and SALL3 in chicken prehierarchical follicle development is unknown. This study aimed to explore the potential role and mechanism of csal1 and csal3 in granulosa cell proliferation, differentiation, and follicle selection within the prehierarchical follicles of hen ovary. Our data demonstrated that the csal1 and csal3 transcriptions were highly expressed in granulosa cells of prehierarchical follicles, and their proteins were mainly localized in the cytoplasm of granulosa cells and oocytes as well as in the ovarian stroma and epithelium. It initially revealed that both csal1 and csal3 may be involved in chicken prehierarchical follicle development via a translocation mechanism. Furthermore, our results showed an abundance of CCND1, Bcat, StAR, CYP11A1, and FSHR mRNA in granulosa cells, and the proliferation levels of granulosa cells from the prehierarchical follicles were significantly increased by siRNA-mediated knockdown of csal1 or/and csal3. Conversely, the overexpression of csal1 or/and csal3 in the granulosa cells led to a remarkably decreased of them. Moreover, csal1 and csal3 together exert a much stronger effect on the regulation than any of csal1 or csal3. These results indicated that csal1 and csal3 play synergistic inhibitory roles on granulosa cell proliferation, differentiation, and steroidogenesis during prehierarchical follicle development in vitro. The current data provide a basis of molecular mechanisms of csal1 and csal3 in controlling the prehierarchical follicle development and growth of hen ovary in vivo.
Keywords: csal1; csal3; differentiation; follicle selection; proliferation; synergistic inhibition.
© The Author(s) 2019. Published by Oxford University Press on behalf of Society for the Study of Reproduction. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Implication of SLIT3-ROBO1/ROBO2 in granulosa cell proliferation, differentiation and follicle selection in the prehierarchical follicles of hen ovary.Cell Biol Int. 2018 Dec;42(12):1643-1657. doi: 10.1002/cbin.11063. Epub 2018 Oct 17. Cell Biol Int. 2018. PMID: 30288875
-
Cooperative Effects of FOXL2 with the Members of TGF-β Superfamily on FSH Receptor mRNA Expression and Granulosa Cell Proliferation from Hen Prehierarchical Follicles.PLoS One. 2015 Oct 23;10(10):e0141062. doi: 10.1371/journal.pone.0141062. eCollection 2015. PLoS One. 2015. PMID: 26496659 Free PMC article.
-
Determination of the roles of GREM1 gene in granulosa cell proliferation and steroidogenesis of hen ovarian prehierarchical follicles.Theriogenology. 2020 Jul 15;151:28-40. doi: 10.1016/j.theriogenology.2020.03.030. Epub 2020 Mar 27. Theriogenology. 2020. PMID: 32251937
-
RNA-seq identifies differentially expressed genes involved in csal1 overexpression in granulosa cells of prehierarchical follicles in Chinese Dagu hens.Poult Sci. 2023 Jan;102(1):102310. doi: 10.1016/j.psj.2022.102310. Epub 2022 Nov 4. Poult Sci. 2023. PMID: 36442307 Free PMC article.
-
Dynamics of avian ovarian follicle development: cellular mechanisms of granulosa cell differentiation.Gen Comp Endocrinol. 2009 Sep 1;163(1-2):12-7. doi: 10.1016/j.ygcen.2008.11.012. Epub 2008 Nov 27. Gen Comp Endocrinol. 2009. PMID: 19059411 Review.
Cited by
-
Effects of RAC1 on Proliferation of Hen Ovarian Prehierarchical Follicle Granulosa Cells.Animals (Basel). 2020 Sep 6;10(9):1589. doi: 10.3390/ani10091589. Animals (Basel). 2020. PMID: 32899947 Free PMC article.
-
Grade follicles transcriptional profiling analysis in different laying stages in chicken.BMC Genomics. 2022 Jul 7;23(1):492. doi: 10.1186/s12864-022-08728-w. BMC Genomics. 2022. PMID: 35794517 Free PMC article.
-
Transcriptome comparative analysis of ovarian follicles reveals the key genes and signaling pathways implicated in hen egg production.BMC Genomics. 2021 Dec 15;22(1):899. doi: 10.1186/s12864-021-08213-w. BMC Genomics. 2021. PMID: 34911438 Free PMC article.
-
Synthesis, Regulatory Factors, and Signaling Pathways of Estrogen in the Ovary.Reprod Sci. 2023 Feb;30(2):350-360. doi: 10.1007/s43032-022-00932-z. Epub 2022 Apr 6. Reprod Sci. 2023. PMID: 35384637 Review.
-
Cellular Processes in Human Ovarian Follicles Are Regulated by Expression Profile of New Gene Markers-Clinical Approach.J Clin Med. 2021 Dec 24;11(1):73. doi: 10.3390/jcm11010073. J Clin Med. 2021. PMID: 35011815 Free PMC article.
References
-
- Gilchrist RB, Ritter LJ, Myllymaa S, Kaivo-Oja N, Dragovic RA, Hickey TE, Ritvos O, Mottershead DG. Molecular basis of oocyte-paracrine signaling that promotes granulosa cell proliferation. J Cell Sci 2006; 119:3811–3821. - PubMed
-
- Qin N, Fan XC, Zhang YY, Xu XX, Tyasi TL, Jing Y, Mu F, Wei ML, Xu RF. New insights into implication of the SLIT/ROBO pathway in the prehierarchical follicle development of hen ovary. Poult Sci 2015; 94:2235–2246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

