Direct observation of branching MT nucleation in living animal cells
- PMID: 31340987
- PMCID: PMC6719462
- DOI: 10.1083/jcb.201904114
Direct observation of branching MT nucleation in living animal cells
Abstract
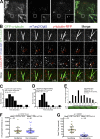
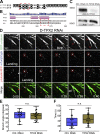
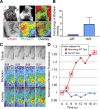
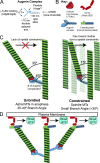
Centrosome-mediated microtubule (MT) nucleation has been well characterized; however, numerous noncentrosomal MT nucleation mechanisms exist. The branching MT nucleation pathway envisages that the γ-tubulin ring complex (γ-TuRC) is recruited to MTs by the augmin complex to initiate nucleation of new MTs. While the pathway is well conserved at a molecular and functional level, branching MT nucleation by core constituents has never been directly observed in animal cells. Here, multicolor TIRF microscopy was applied to visualize and quantitatively define the entire process of branching MT nucleation in dividing Drosophila cells during anaphase. The steps of a stereotypical branching nucleation event entailed augmin binding to a mother MT and recruitment of γ-TuRC after 15 s, followed by nucleation 16 s later of a daughter MT at a 36° branch angle. Daughters typically remained attached throughout their ∼40-s lifetime unless the mother depolymerized past the branch point. Assembly of branched MT arrays, which did not require Drosophila TPX2, enhanced localized RhoA activation during cytokinesis.
© 2019 Verma and Maresca.
Figures

Similar articles
-
In vitro reconstitution of branching microtubule nucleation.Elife. 2020 Jan 14;9:e49769. doi: 10.7554/eLife.49769. Elife. 2020. PMID: 31933481 Free PMC article.
-
Structural analysis of the role of TPX2 in branching microtubule nucleation.J Cell Biol. 2017 Apr 3;216(4):983-997. doi: 10.1083/jcb.201607060. Epub 2017 Mar 6. J Cell Biol. 2017. PMID: 28264915 Free PMC article.
-
How Microtubules Build the Spindle Branch by Branch.Annu Rev Cell Dev Biol. 2022 Oct 6;38:1-23. doi: 10.1146/annurev-cellbio-120420-114559. Epub 2022 Jun 27. Annu Rev Cell Dev Biol. 2022. PMID: 35759800 Free PMC article. Review.
-
Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells.New Phytol. 2019 Jun;222(4):1705-1718. doi: 10.1111/nph.15705. Epub 2019 Feb 25. New Phytol. 2019. PMID: 30681146 Review.
-
Spatiotemporal organization of branched microtubule networks.Elife. 2019 May 8;8:e43890. doi: 10.7554/eLife.43890. Elife. 2019. PMID: 31066674 Free PMC article.
Cited by
-
The model of local axon homeostasis - explaining the role and regulation of microtubule bundles in axon maintenance and pathology.Neural Dev. 2019 Nov 9;14(1):11. doi: 10.1186/s13064-019-0134-0. Neural Dev. 2019. PMID: 31706327 Free PMC article. Review.
-
A celebration of the 25th anniversary of chromatin-mediated spindle assembly.Mol Biol Cell. 2022 Feb 1;33(2):rt1. doi: 10.1091/mbc.E21-08-0400. Mol Biol Cell. 2022. PMID: 35076260 Free PMC article.
-
Augmin complex activity finetunes dendrite morphology through non-centrosomal microtubule nucleation in vivo.J Cell Sci. 2024 May 1;137(9):jcs261512. doi: 10.1242/jcs.261512. Epub 2024 May 10. J Cell Sci. 2024. PMID: 38587100 Free PMC article.
-
γ-TuRCs and the augmin complex are required for the development of highly branched dendritic arbors in Drosophila.J Cell Sci. 2024 May 1;137(9):jcs261534. doi: 10.1242/jcs.261534. Epub 2024 May 10. J Cell Sci. 2024. PMID: 38606636 Free PMC article.
-
Biochemical reconstitution of branching microtubule nucleation.Elife. 2020 Jan 14;9:e49797. doi: 10.7554/eLife.49797. Elife. 2020. PMID: 31933480 Free PMC article.
References
-
- Basnet N., Nedozralova H., Crevenna A.H., Bodakuntla S., Schlichthaerle T., Taschner M., Cardone G., Janke C., Jungmann R., Magiera M.M., et al. . 2018. Direct induction of microtubule branching by microtubule nucleation factor SSNA1. Nat. Cell Biol. 20:1172–1180. 10.1038/s41556-018-0199-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

