Simulated microgravity increases polyploid giant cancer cells and nuclear localization of YAP
- PMID: 31337825
- PMCID: PMC6650394
- DOI: 10.1038/s41598-019-47116-5
Simulated microgravity increases polyploid giant cancer cells and nuclear localization of YAP
Abstract
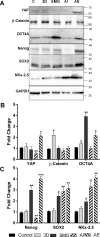
Physical cues are vital in determining cellular fate in cancer. In vitro 3D culture do not replicate forces present in vivo. These forces including tumor interstitial fluid pressure and matrix stiffness behave as switches in differentiation and metastasis, which are intricate features of cancer stem cells (CSCs). Gravity determines the effect of these physical factors on cell fate and functions as evident from microgravity experiments on space and ground simulations. Here, we described the role of simulation of microgravity (SMG) using rotary cell culture system (RCCS) in increasing stemness in human colorectal cancer cell HCT116. We observed distinct features of cancer stem cells including CD133/CD44 dual positive cells and migration in SMG which was not altered by autophagy induction or inhibition. 3D and SMG increased autophagy, but the flux was staggered under SMG. Increased unique giant cancer cells housing complete nuclear localization of YAP were observed in SMG. This study highlights the role of microgravity in regulating stemness in CSC and importance of physical factors in determining the same.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
IL-8/CXCR2 mediates tropism of human bone marrow-derived mesenchymal stem cells toward CD133+ /CD44+ Colon cancer stem cells.J Cell Physiol. 2021 Apr;236(4):3114-3128. doi: 10.1002/jcp.30080. Epub 2020 Oct 20. J Cell Physiol. 2021. PMID: 33078417
-
CD44+ and CD133+ Non-Small Cell Lung Cancer Cells Exhibit DNA Damage Response Pathways and Dormant Polyploid Giant Cancer Cell Enrichment Relating to Their p53 Status.Int J Mol Sci. 2022 Apr 28;23(9):4922. doi: 10.3390/ijms23094922. Int J Mol Sci. 2022. PMID: 35563313 Free PMC article.
-
A rational approach for cancer stem-like cell isolation and characterization using CD44 and prominin-1(CD133) as selection markers.Oncotarget. 2016 Nov 29;7(48):78499-78515. doi: 10.18632/oncotarget.12100. Oncotarget. 2016. PMID: 27655682 Free PMC article.
-
Hyaluronan-CD44 axis orchestrates cancer stem cell functions.Cell Signal. 2019 Nov;63:109377. doi: 10.1016/j.cellsig.2019.109377. Epub 2019 Jul 27. Cell Signal. 2019. PMID: 31362044 Review.
-
Clinicopathological and prognostic significance of cancer stem cell markers CD44 and CD133 in patients with gastric cancer: A comprehensive meta-analysis with 4729 patients involved.Medicine (Baltimore). 2016 Oct;95(42):e5163. doi: 10.1097/MD.0000000000005163. Medicine (Baltimore). 2016. PMID: 27759647 Free PMC article. Review.
Cited by
-
High Migration and Invasion Ability of PGCCs and Their Daughter Cells Associated With the Nuclear Localization of S100A10 Modified by SUMOylation.Front Cell Dev Biol. 2021 Jul 16;9:696871. doi: 10.3389/fcell.2021.696871. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34336846 Free PMC article.
-
Omics Studies of Tumor Cells under Microgravity Conditions.Int J Mol Sci. 2024 Jan 11;25(2):926. doi: 10.3390/ijms25020926. Int J Mol Sci. 2024. PMID: 38255998 Free PMC article. Review.
-
Exosomes derived from umbilical cord mesenchymal stem cells in mechanical environment show improved osteochondral activity via upregulation of LncRNA H19.J Orthop Translat. 2020 Apr 3;26:111-120. doi: 10.1016/j.jot.2020.03.005. eCollection 2021 Jan. J Orthop Translat. 2020. PMID: 33437630 Free PMC article.
-
The effects of microgravity on differentiation and cell growth in stem cells and cancer stem cells.Stem Cells Transl Med. 2020 Aug;9(8):882-894. doi: 10.1002/sctm.20-0084. Epub 2020 Apr 30. Stem Cells Transl Med. 2020. PMID: 32352658 Free PMC article.
-
5FU encapsulated polyglycerol sebacate nanoparticles as anti-cancer drug carriers.RSC Adv. 2021 May 25;11(31):18984-18993. doi: 10.1039/d1ra01722e. eCollection 2021 May 24. RSC Adv. 2021. PMID: 35478658 Free PMC article.
References
-
- Ariffin A, Forde P, Jahangeer S, Soden D, Hinchion J. Releasing Pressure in Tumors: What Do We Know So Far and Where Do We Go from Here? A Review. Cancer Research. 2014;74:2655–2662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

