Identification of Dephospho-Coenzyme A (Dephospho-CoA) Kinase in Thermococcus kodakarensis and Elucidation of the Entire CoA Biosynthesis Pathway in Archaea
- PMID: 31337720
- PMCID: PMC6650551
- DOI: 10.1128/mBio.01146-19
Identification of Dephospho-Coenzyme A (Dephospho-CoA) Kinase in Thermococcus kodakarensis and Elucidation of the Entire CoA Biosynthesis Pathway in Archaea
Abstract
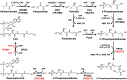
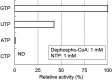
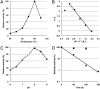
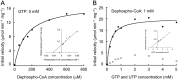
Dephospho-coenzyme A (dephospho-CoA) kinase (DPCK) catalyzes the ATP-dependent phosphorylation of dephospho-CoA, the final step in coenzyme A (CoA) biosynthesis. DPCK has been identified and characterized in bacteria and eukaryotes but not in archaea. The hyperthermophilic archaeon Thermococcus kodakarensis encodes two homologs of bacterial DPCK and the DPCK domain of eukaryotic CoA synthase, TK1334 and TK2192. We purified the recombinant TK1334 and TK2192 proteins and found that they lacked DPCK activity. Bioinformatic analyses showed that, in several archaea, the uncharacterized gene from arCOG04076 protein is fused with the gene for phosphopantetheine adenylyltransferase (PPAT), which catalyzes the reaction upstream of the DPCK reaction in CoA biosynthesis. This observation suggested that members of arCOG04076, both fused to PPAT and standalone, could be the missing archaeal DPCKs. We purified the recombinant TK1697 protein, a standalone member of arCOG04076 from T. kodakarensis, and demonstrated its GTP-dependent DPCK activity. Disruption of the TK1697 resulted in CoA auxotrophy, indicating that TK1697 encodes a DPCK that contributes to CoA biosynthesis in T. kodakarensis TK1697 homologs are widely distributed in archaea, suggesting that the arCOG04076 protein represents a novel family of DPCK that is not homologous to bacterial and eukaryotic DPCKs but is distantly related to bacterial and eukaryotic thiamine pyrophosphokinases. We also constructed and characterized gene disruption strains of TK0517 and TK2128, homologs of bifunctional phosphopantothenoylcysteine synthetase-phosphopantothenoylcysteine decarboxylase and PPAT, respectively. Both strains displayed CoA auxotrophy, indicating their contribution to CoA biosynthesis. Taken together with previous studies, the results experimentally validate the entire CoA biosynthesis pathway in T. kodakarensisIMPORTANCE CoA is utilized in a wide range of metabolic pathways, and its biosynthesis is essential for all life. Pathways for CoA biosynthesis in bacteria and eukaryotes have been established. In archaea, however, the enzyme that catalyzes the final step in CoA biosynthesis, dephospho-CoA kinase (DPCK), had not been identified. In the present study, bioinformatic analyses identified a candidate for the DPCK in archaea, which was biochemically and genetically confirmed in the hyperthermophilic archaeon Thermococcus kodakarensis Genetic analyses on genes presumed to encode bifunctional phosphopantothenoylcysteine synthetase-phosphopantothenoylcysteine decarboxylase and phosphopantetheine adenylyltransferase confirmed their involvement in CoA biosynthesis. Taken together with previous studies, the results reveal the entire pathway for CoA biosynthesis in a single archaeon and provide insight into the different mechanisms of CoA biosynthesis and their distribution in nature.
Keywords: archaea; coenzyme A; dephospho-CoA kinase; hyperthermophiles; metabolism.
Copyright © 2019 Shimosaka et al.
Figures
Similar articles
-
Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.Proteins. 2024 Jun;92(6):768-775. doi: 10.1002/prot.26666. Epub 2024 Jan 18. Proteins. 2024. PMID: 38235908
-
Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea.J Biol Chem. 2009 Oct 9;284(41):28137-28145. doi: 10.1074/jbc.M109.009696. Epub 2009 Aug 7. J Biol Chem. 2009. PMID: 19666462 Free PMC article.
-
A detailed biochemical characterization of phosphopantothenate synthetase, a novel enzyme involved in coenzyme A biosynthesis in the Archaea.Extremophiles. 2012 Nov;16(6):819-28. doi: 10.1007/s00792-012-0477-5. Epub 2012 Sep 2. Extremophiles. 2012. PMID: 22940806
-
CoA biosynthesis in archaea.Biochem Soc Trans. 2013 Feb 1;41(1):427-31. doi: 10.1042/BST20120311. Biochem Soc Trans. 2013. PMID: 23356323 Review.
-
Phosphopantetheine Adenylyltransferase: A promising drug target to combat antibiotic resistance.Biochim Biophys Acta Proteins Proteom. 2021 Feb;1869(2):140566. doi: 10.1016/j.bbapap.2020.140566. Epub 2020 Nov 30. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 33271445 Review.
Cited by
-
Integration of large heterologous DNA fragments into the genome of Thermococcus kodakarensis.Extremophiles. 2020 May;24(3):339-353. doi: 10.1007/s00792-020-01159-z. Epub 2020 Feb 28. Extremophiles. 2020. PMID: 32112303
-
Vanin 1 Gene Role in Modulation of iNOS/MCP-1/TGF-β1 Signaling Pathway in Obese Diabetic Patients.J Inflamm Res. 2022 Dec 14;15:6745-6759. doi: 10.2147/JIR.S386506. eCollection 2022. J Inflamm Res. 2022. PMID: 36540060 Free PMC article.
-
In Vitro Production of Coenzyme A Using Thermophilic Enzymes.Appl Environ Microbiol. 2021 Jun 25;87(14):e0054121. doi: 10.1128/AEM.00541-21. Epub 2021 Jun 25. Appl Environ Microbiol. 2021. PMID: 33990309 Free PMC article.
-
The TK0271 Protein Activates Transcription of Aromatic Amino Acid Biosynthesis Genes in the Hyperthermophilic Archaeon Thermococcus kodakarensis.mBio. 2019 Sep 10;10(5):e01213-19. doi: 10.1128/mBio.01213-19. mBio. 2019. PMID: 31506306 Free PMC article.
-
A New Species-Specific Typing Method for Salivarius Group Streptococci Based on the Dephospho-Coenzyme A Kinase (coaE) Gene Sequencing.Front Cell Infect Microbiol. 2021 Aug 6;11:685657. doi: 10.3389/fcimb.2021.685657. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34422679 Free PMC article.
References
-
- Ishibashi T, Tomita H, Yokooji Y, Morikita T, Watanabe B, Hiratake J, Kishimoto A, Kita A, Miki K, Imanaka T, Atomi H. 2012. A detailed biochemical characterization of phosphopantothenate synthetase, a novel enzyme involved in coenzyme A biosynthesis in the Archaea. Extremophiles 16:819–828. doi:10.1007/s00792-012-0477-5. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
