The Cajal Body Protein WRAP53β Prepares the Scene for Repair of DNA Double-Strand Breaks by Regulating Local Ubiquitination
- PMID: 31334247
- PMCID: PMC6624377
- DOI: 10.3389/fmolb.2019.00051
The Cajal Body Protein WRAP53β Prepares the Scene for Repair of DNA Double-Strand Breaks by Regulating Local Ubiquitination
Abstract
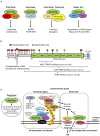
Proper repair of DNA double-strand breaks is critical for maintaining genome integrity and avoiding disease. Modification of damaged chromatin has profound consequences for the initial signaling and regulation of repair. One such modification involves ubiquitination by E3 ligases RNF8 and RNF168 within minutes after DNA double-strand break formation, altering chromatin structure and recruiting factors such as 53BP1 and BRCA1 for repair via non-homologous end-joining (NHEJ) and homologous recombination (HR), respectively. The WD40 protein WRAP53β plays an essential role in localizing RNF8 to DNA breaks by scaffolding its interaction with the upstream factor MDC1. Loss of WRAP53β impairs ubiquitination at DNA lesions and reduces downstream repair by both NHEJ and HR. Intriguingly, WRAP53β depletion attenuates repair of DNA double-strand breaks more than depletion of RNF8, indicating functions other than RNF8-mediated ubiquitination. WRAP53β plays key roles with respect to the nuclear organelles Cajal bodies, including organizing the genome to promote associated transcription and collecting factors involved in maturation of the spliceosome and telomere elongation within these organelles. It is possible that similar functions may aid also in DNA repair. Here we describe the involvement of WRAP53β in Cajal bodies and DNA double-strand break repair in detail and explore whether and how these processes may be linked. We also discuss the possibility that the overexpression of WRAP53β detected in several cancer types may reflect its normal participation in the DNA damage response rather than oncogenic properties.
Keywords: Cajal body; DNA repair; RNF8; WD40; WRAP53β; cancer; chromatin modification; ubiquitin.
Figures
Similar articles
-
Overexpression of the scaffold WD40 protein WRAP53β enhances the repair of and cell survival from DNA double-strand breaks.Cell Death Dis. 2016 Jun 16;7(6):e2267. doi: 10.1038/cddis.2016.172. Cell Death Dis. 2016. PMID: 27310875 Free PMC article.
-
The scaffold protein WRAP53β orchestrates the ubiquitin response critical for DNA double-strand break repair.Genes Dev. 2014 Dec 15;28(24):2726-38. doi: 10.1101/gad.246546.114. Genes Dev. 2014. PMID: 25512560 Free PMC article.
-
Phosphorylation of the Cajal body protein WRAP53β by ATM promotes its involvement in the DNA damage response.RNA Biol. 2017 Jun 3;14(6):804-813. doi: 10.1080/15476286.2016.1243647. Epub 2016 Oct 7. RNA Biol. 2017. PMID: 27715493 Free PMC article.
-
Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice.J Radiat Res. 2016 Aug;57 Suppl 1(Suppl 1):i33-i40. doi: 10.1093/jrr/rrw027. Epub 2016 Mar 16. J Radiat Res. 2016. PMID: 26983989 Free PMC article. Review.
-
On the road with WRAP53β: guardian of Cajal bodies and genome integrity.Front Genet. 2015 Mar 24;6:91. doi: 10.3389/fgene.2015.00091. eCollection 2015. Front Genet. 2015. PMID: 25852739 Free PMC article. Review.
Cited by
-
Evaluation of dyskerin expression and the Cajal body protein WRAP53β as potential prognostic markers for patients with primary vaginal carcinoma.Oncol Lett. 2022 Jan;23(1):30. doi: 10.3892/ol.2021.13148. Epub 2021 Nov 23. Oncol Lett. 2022. PMID: 34868367 Free PMC article.
-
The Nucleolus and Its Interactions with Viral Proteins Required for Successful Infection.Cells. 2024 Sep 21;13(18):1591. doi: 10.3390/cells13181591. Cells. 2024. PMID: 39329772 Free PMC article. Review.
-
Biallelic mutations in WRAP53 result in dysfunctional telomeres, Cajal bodies and DNA repair, thereby causing Hoyeraal-Hreidarsson syndrome.Cell Death Dis. 2020 Apr 17;11(4):238. doi: 10.1038/s41419-020-2421-4. Cell Death Dis. 2020. PMID: 32303682 Free PMC article.
-
Low levels of WRAP53 predict decreased efficacy of radiotherapy and are prognostic for local recurrence and death from breast cancer: a long-term follow-up of the SweBCG91RT randomized trial.Mol Oncol. 2023 Oct;17(10):2029-2040. doi: 10.1002/1878-0261.13426. Epub 2023 Apr 19. Mol Oncol. 2023. PMID: 36975842 Free PMC article. Clinical Trial.
-
Validation and Analysis of COIL, a Gene Associated with Multiple Lambing Traits in Sheep.Genes (Basel). 2024 Feb 13;15(2):235. doi: 10.3390/genes15020235. Genes (Basel). 2024. PMID: 38397224 Free PMC article.
References
-
- Baranes-Bachar K., Levy-Barda A., Oehler J., Reid D. A., Soria-Bretones I., Voss T. C., et al. . (2018). The ubiquitin E3/E4 ligase UBE4A adjusts protein ubiquitylation and accumulation at sites of DNA damage, facilitating double-strand break repair. Mol. Cell 69, 866–878.e867. 10.1016/j.molcel.2018.02.002 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

