Copine A Interacts with Actin Filaments and Plays a Role in Chemotaxis and Adhesion
- PMID: 31330887
- PMCID: PMC6679068
- DOI: 10.3390/cells8070758
Copine A Interacts with Actin Filaments and Plays a Role in Chemotaxis and Adhesion
Abstract
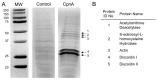
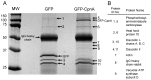
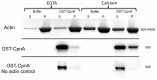
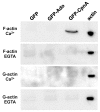
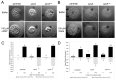
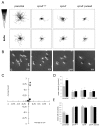
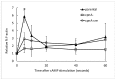
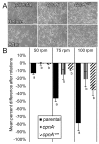
Copines make up a family of calcium-dependent, phospholipid-binding proteins found in numerous eukaryotic organisms. Copine proteins consist of two C2 domains at the N-terminus followed by an A domain similar to the von Willebrand A domain found in integrins. We are studying copine protein function in the model organism, Dictyostelium discoideum, which has six copine genes, cpnA-cpnF. Previous research showed that cells lacking the cpnA gene exhibited a cytokinesis defect, a contractile vacuole defect, and developmental defects. To provide insight into the role of CpnA in these cellular processes, we used column chromatography and immunoprecipitation to isolate proteins that bind to CpnA. These proteins were identified by mass spectrometry. One of the proteins identified was actin. Purified CpnA was shown to bind to actin filaments in a calcium-dependent manner in vitro. cpnA- cells exhibited defects in three actin-based processes: chemotaxis, cell polarity, and adhesion. These results suggest that CpnA plays a role in chemotaxis and adhesion and may do so by interacting with actin filaments.
Keywords: Dictyostelium; actin; adhesion; cAMP; calcium; chemotaxis; copine.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Copine A regulates the size and exocytosis of contractile vacuoles and postlysosomes in Dictyostelium.FEBS Open Bio. 2020 Jun;10(6):979-994. doi: 10.1002/2211-5463.12874. Epub 2020 May 19. FEBS Open Bio. 2020. PMID: 32351039 Free PMC article.
-
Cyclic AMP signaling in Dictyostelium promotes the translocation of the copine family of calcium-binding proteins to the plasma membrane.BMC Cell Biol. 2018 Jul 16;19(1):13. doi: 10.1186/s12860-018-0160-5. BMC Cell Biol. 2018. PMID: 30012091 Free PMC article.
-
Copine A, a calcium-dependent membrane-binding protein, transiently localizes to the plasma membrane and intracellular vacuoles in Dictyostelium.BMC Cell Biol. 2005 Dec 12;6:46. doi: 10.1186/1471-2121-6-46. BMC Cell Biol. 2005. PMID: 16343335 Free PMC article.
-
Cell migration: regulation of cytoskeleton by Rap1 in Dictyostelium discoideum.J Microbiol. 2012 Aug;50(4):555-61. doi: 10.1007/s12275-012-2246-7. Epub 2012 Aug 25. J Microbiol. 2012. PMID: 22923101 Review.
-
Cell-cell signaling and adhesion in phagocytosis and early development of Dictyostelium.Int J Dev Biol. 2000;44(6):733-42. Int J Dev Biol. 2000. PMID: 11061438 Review.
Cited by
-
Copine A regulates the size and exocytosis of contractile vacuoles and postlysosomes in Dictyostelium.FEBS Open Bio. 2020 Jun;10(6):979-994. doi: 10.1002/2211-5463.12874. Epub 2020 May 19. FEBS Open Bio. 2020. PMID: 32351039 Free PMC article.
-
Copine C plays a role in adhesion and streaming in Dictyostelium.Cell Adh Migr. 2024 Dec;18(1):1-19. doi: 10.1080/19336918.2024.2315629. Epub 2024 Feb 20. Cell Adh Migr. 2024. PMID: 38378453 Free PMC article.
-
Adhesion of Dictyostelium Amoebae to Surfaces: A Brief History of Attachments.Front Cell Dev Biol. 2022 May 27;10:910736. doi: 10.3389/fcell.2022.910736. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35721508 Free PMC article. Review.
-
Molecular studies into cell biological role of Copine-4 in Retinal Ganglion Cells.PLoS One. 2021 Nov 30;16(11):e0255860. doi: 10.1371/journal.pone.0255860. eCollection 2021. PLoS One. 2021. PMID: 34847148 Free PMC article.
-
Phosphatidylserine exposure promotes increased adhesion in Dictyostelium Copine A mutants.PLoS One. 2021 May 27;16(5):e0250710. doi: 10.1371/journal.pone.0250710. eCollection 2021. PLoS One. 2021. PMID: 34043641 Free PMC article.
References
-
- Creutz C.E., Tomsig J.L., Snyder S.L., Gautier M.C., Skouri F., Beisson J., Cohen J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 1998;273:1393–1402. doi: 10.1074/jbc.273.3.1393. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

