Mutually exclusive amino acid residues of L13a are responsible for its ribosomal incorporation and translational silencing leading to resolution of inflammation
- PMID: 31308261
- PMCID: PMC6800476
- DOI: 10.1261/rna.071118.119
Mutually exclusive amino acid residues of L13a are responsible for its ribosomal incorporation and translational silencing leading to resolution of inflammation
Abstract
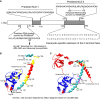
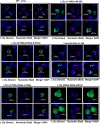
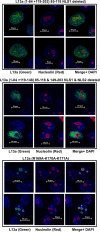
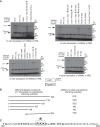
Eukaryotic ribosomal protein L13a is a member of the conserved universal ribosomal uL13 protein family. Structurally, L13a is distinguished from its prokaryotic counterparts by the presence of an ∼55 amino acid-long carboxy-terminal α-helical extension. The importance of these evolved residues in the carboxy-terminal extension for mammalian ribosome biogenesis as well as L13a's extraribosomal function in GAIT (γ interferon-activated inhibitor of translation) complex-mediated translation silencing during inflammation is not understood. Here, we present biochemical analyses of L13a mutant variants identifying several mutually exclusive amino acid residues in the eukaryote-specific carboxy-terminal extension of human L13a (Tyr149-Val203) important for ribosomal incorporation and translational silencing. Specifically, we show that mutation of Arg169, Lys170, and Lys171 to Ala abrogate GAIT-mediated translational silencing, but not L13a incorporation into ribosomes. Moreover, we show that the carboxy-terminal helix alone can silence translation of GAIT element-containing mRNAs in vitro. We also show through cellular immunofluorescence experiments that nuclear but not nucleolar localization of L13a is resistant to extensive amino acid alterations, suggesting that multiple complex nuclear import signals are present within this protein. These studies provide new insights into L13a structure and its ribosomal and extraribosomal functions in model human cells.
Keywords: L13a; inflammation; nuclear localization; ribosomal incorporation; translational silencing.
© 2019 Kour et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



Similar articles
-
Insights into the mechanism of ribosomal incorporation of mammalian L13a protein during ribosome biogenesis.Mol Cell Biol. 2013 Aug;33(15):2829-42. doi: 10.1128/MCB.00250-13. Epub 2013 May 20. Mol Cell Biol. 2013. PMID: 23689135 Free PMC article.
-
Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control.Cell. 2003 Oct 17;115(2):187-98. doi: 10.1016/s0092-8674(03)00773-6. Cell. 2003. PMID: 14567916
-
Extraribosomal l13a is a specific innate immune factor for antiviral defense.J Virol. 2014 Aug;88(16):9100-10. doi: 10.1128/JVI.01129-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899178 Free PMC article.
-
The GAIT translational control system.Wiley Interdiscip Rev RNA. 2018 Mar;9(2):e1441. doi: 10.1002/wrna.1441. Epub 2017 Nov 20. Wiley Interdiscip Rev RNA. 2018. PMID: 29152905 Free PMC article. Review.
-
Eukaryotic protein uS19: a component of the decoding site of ribosomes and a player in human diseases.Biochem J. 2021 Mar 12;478(5):997-1008. doi: 10.1042/BCJ20200950. Biochem J. 2021. PMID: 33661277 Review.
Cited by
-
Validation of reference genes for the normalization of the RT-qPCR in peripheral blood mononuclear cells of septic patients.Heliyon. 2023 Apr 7;9(4):e15269. doi: 10.1016/j.heliyon.2023.e15269. eCollection 2023 Apr. Heliyon. 2023. PMID: 37089378 Free PMC article.
References
-
- Basu A, Poddar D, Robinet P, Smith JD, Febbraio M, Baldwin WM III, Mazumder B. 2014. Ribosomal protein L13a deficiency in macrophages promotes atherosclerosis by limiting translation control-dependent retardation of inflammation. Arterioscler Thromb Vasc Biol 34: 533–542. 10.1161/ATVBAHA.113.302573 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
