Non-Coding RNAs and their Integrated Networks
- PMID: 31301674
- PMCID: PMC6798851
- DOI: 10.1515/jib-2019-0027
Non-Coding RNAs and their Integrated Networks
Abstract
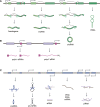
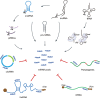
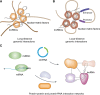
Eukaryotic genomes are pervasively transcribed. Besides protein-coding RNAs, there are different types of non-coding RNAs that modulate complex molecular and cellular processes. RNA sequencing technologies and bioinformatics methods greatly promoted the study of ncRNAs, which revealed ncRNAs' essential roles in diverse aspects of biological functions. As important key players in gene regulatory networks, ncRNAs work with other biomolecules, including coding and non-coding RNAs, DNAs and proteins. In this review, we discuss the distinct types of ncRNAs, including housekeeping ncRNAs and regulatory ncRNAs, their versatile functions and interactions, transcription, translation, and modification. Moreover, we summarize the integrated networks of ncRNA interactions, providing a comprehensive landscape of ncRNAs regulatory roles.
Keywords: ceRNA network; integrated network; ncRNA interaction; non-coding RNA; regulatory ncRNA.
Conflict of interest statement
Authors state no conflict of interest. All authors have read the journal’s publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
Figures
Comment in
-
Editorial: Role of long non-coding RNA and Circular RNA in bone metabolism and their role as circulating biomarkers for bone diseases.Front Endocrinol (Lausanne). 2023 Nov 14;14:1321962. doi: 10.3389/fendo.2023.1321962. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 38034002 Free PMC article. No abstract available.
Similar articles
-
Advances in Computational Methodologies for Classification and Sub-Cellular Locality Prediction of Non-Coding RNAs.Int J Mol Sci. 2021 Aug 13;22(16):8719. doi: 10.3390/ijms22168719. Int J Mol Sci. 2021. PMID: 34445436 Free PMC article. Review.
-
Biogenesis, Functions, Interactions, and Resources of Non-Coding RNAs in Plants.Int J Mol Sci. 2022 Mar 28;23(7):3695. doi: 10.3390/ijms23073695. Int J Mol Sci. 2022. PMID: 35409060 Free PMC article. Review.
-
Versatile interactions and bioinformatics analysis of noncoding RNAs.Brief Bioinform. 2019 Sep 27;20(5):1781-1794. doi: 10.1093/bib/bby050. Brief Bioinform. 2019. PMID: 29939215 Review.
-
Non-Coding RNAs in Tuberculosis Epidemiology: Platforms and Approaches for Investigating the Genome's Dark Matter.Int J Mol Sci. 2022 Apr 17;23(8):4430. doi: 10.3390/ijms23084430. Int J Mol Sci. 2022. PMID: 35457250 Free PMC article. Review.
-
Emerging Classes of Small Non-Coding RNAs With Potential Implications in Diabetes and Associated Metabolic Disorders.Front Endocrinol (Lausanne). 2021 May 10;12:670719. doi: 10.3389/fendo.2021.670719. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34040585 Free PMC article. Review.
Cited by
-
Non-Coding RNAs and Innate Immune Responses in Cancer.Biomedicines. 2024 Sep 11;12(9):2072. doi: 10.3390/biomedicines12092072. Biomedicines. 2024. PMID: 39335585 Free PMC article. Review.
-
Building a Human Ovarian Antioxidant ceRNA Network "OvAnOx": A Bioinformatic Perspective for Research on Redox-Related Ovarian Functions and Dysfunctions.Antioxidants (Basel). 2024 Sep 12;13(9):1101. doi: 10.3390/antiox13091101. Antioxidants (Basel). 2024. PMID: 39334761 Free PMC article. Review.
-
The regulatory landscape of interacting RNA and protein pools in cellular homeostasis and cancer.Hum Genomics. 2024 Sep 27;18(1):109. doi: 10.1186/s40246-024-00678-6. Hum Genomics. 2024. PMID: 39334294 Free PMC article. Review.
-
Epigenetic mechanisms in cardiovascular complications of diabetes: towards future therapies.Mol Med. 2024 Sep 27;30(1):161. doi: 10.1186/s10020-024-00939-z. Mol Med. 2024. PMID: 39333854 Free PMC article. Review.
-
The Nucleolus and Its Interactions with Viral Proteins Required for Successful Infection.Cells. 2024 Sep 21;13(18):1591. doi: 10.3390/cells13181591. Cells. 2024. PMID: 39329772 Free PMC article. Review.
References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith M, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559–63. - PubMed
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith M, Maeda N. et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
