Plant Antiviral Immunity Against Geminiviruses and Viral Counter-Defense for Survival
- PMID: 31297106
- PMCID: PMC6607972
- DOI: 10.3389/fmicb.2019.01460
Plant Antiviral Immunity Against Geminiviruses and Viral Counter-Defense for Survival
Abstract
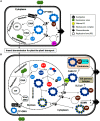
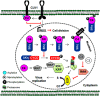
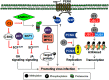
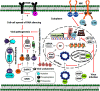
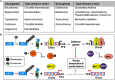
The family Geminiviridae includes plant-infecting viruses whose genomes are composed of one or two circular non-enveloped ssDNAs(+) of about 2.5-5.2 kb each in size. These insect-transmissible geminiviruses cause significant crop losses across continents and pose a serious threat to food security. Under the control of promoters generally located within the intergenic region, their genomes encode five to eight ORFs from overlapping viral transcripts. Most proteins encoded by geminiviruses perform multiple functions, such as suppressing defense responses, hijacking ubiquitin-proteasomal pathways, altering hormonal responses, manipulating cell cycle regulation, and exploiting protein-signaling cascades. Geminiviruses establish complex but coordinated interactions with several host elements to spread and facilitate successful infection cycles. Consequently, plants have evolved several multilayered defense strategies against geminivirus infection and distribution. Recent studies on the evasion of host-mediated resistance factors by various geminivirus proteins through novel mechanisms have provided new insights into the development of antiviral strategies against geminiviruses. This review summarizes the current knowledge concerning virus movement within and between cells, as well as the recent advances in our understanding of the biological roles of virus-encoded proteins in manipulating host-mediated responses and insect transmission. This review also highlights unexplored areas that may increase our understanding of the biology of geminiviruses and how to combat these important plant pathogens.
Keywords: antiviral response; hormone signaling; macromolecular trafficking; post-translational modification; vector transmission; viral pathogenesis; virus.
Figures

Similar articles
-
Plant-virus-insect tritrophic interactions: insights into the functions of geminivirus virion-sense strand genes.Proc Biol Sci. 2020 Oct 14;287(1936):20201846. doi: 10.1098/rspb.2020.1846. Epub 2020 Oct 14. Proc Biol Sci. 2020. PMID: 33049166 Free PMC article.
-
Plant Defense and Viral Counter-Defense during Plant-Geminivirus Interactions.Viruses. 2023 Feb 12;15(2):510. doi: 10.3390/v15020510. Viruses. 2023. PMID: 36851725 Free PMC article. Review.
-
Plant responses to geminivirus infection: guardians of the plant immunity.Virol J. 2021 Jul 9;18(1):143. doi: 10.1186/s12985-021-01612-1. Virol J. 2021. PMID: 34243802 Free PMC article. Review.
-
iTRAQ analysis of the tobacco leaf proteome reveals that RNA-directed DNA methylation (RdDM) has important roles in defense against geminivirus-betasatellite infection.J Proteomics. 2017 Jan 30;152:88-101. doi: 10.1016/j.jprot.2016.10.015. Epub 2016 Oct 29. J Proteomics. 2017. PMID: 27989946
-
Insights into the multifunctional roles of geminivirus-encoded proteins in pathogenesis.Arch Virol. 2022 Feb;167(2):307-326. doi: 10.1007/s00705-021-05338-x. Epub 2022 Jan 26. Arch Virol. 2022. PMID: 35079902 Review.
Cited by
-
Clustered Regularly Interspaced Short Palindromic Repeats-Associated Protein System for Resistance Against Plant Viruses: Applications and Perspectives.Front Plant Sci. 2022 May 26;13:904829. doi: 10.3389/fpls.2022.904829. eCollection 2022. Front Plant Sci. 2022. PMID: 35693174 Free PMC article. Review.
-
Development of a triple antibody sandwich enzyme-linked immunosorbent assay for cassava mosaic disease detection using a monoclonal antibody to Sri Lankan cassava mosaic virus.Virol J. 2021 May 18;18(1):100. doi: 10.1186/s12985-021-01572-6. Virol J. 2021. PMID: 34006310 Free PMC article.
-
Differential expression of genes during recovery of Nicotiana tabacum from tomato leaf curl Gujarat virus infection.Planta. 2023 Jul 5;258(2):37. doi: 10.1007/s00425-023-04182-4. Planta. 2023. PMID: 37405593 Free PMC article.
-
A cassava protoplast system for screening genes associated with the response to South African cassava mosaic virus.Virol J. 2020 Nov 23;17(1):184. doi: 10.1186/s12985-020-01453-4. Virol J. 2020. PMID: 33228712 Free PMC article.
-
Begomovirus-Host Interactions: Viral Proteins Orchestrating Intra and Intercellular Transport of Viral DNA While Suppressing Host Defense Mechanisms.Viruses. 2023 Jul 21;15(7):1593. doi: 10.3390/v15071593. Viruses. 2023. PMID: 37515277 Free PMC article. Review.
References
-
- Amin I., Patil B. L., Briddon R. W., Mansoor S., Fauquet C. M. (2011). Comparison of phenotypes produced in response to transient expression of genes encoded by four distinct begomoviruses in Nicotiana benthamiana and their correlation with the levels of developmental miRNAs. Virol. J. 8:238. 10.1186/1743-422X-8-238 - DOI - PMC - PubMed
-
- Ascencio-Ibáñez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 436–454. 10.1104/pp.108.121038 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

