Functional Enrichment and Analysis of Antigen-Specific Memory B Cell Antibody Repertoires in PBMCs
- PMID: 31293598
- PMCID: PMC6603168
- DOI: 10.3389/fimmu.2019.01452
Functional Enrichment and Analysis of Antigen-Specific Memory B Cell Antibody Repertoires in PBMCs
Abstract
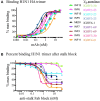
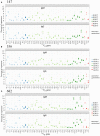
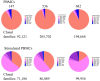
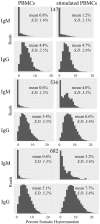
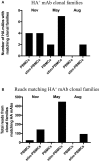
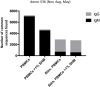
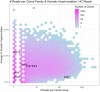
Phenotypic screening of antigen-specific antibodies in human blood is a common diagnostic test for infectious agents and a correlate of protection after vaccination. In addition to long-lived antibody secreting plasma cells residing in the bone marrow, memory B cells are a latent source of antigen-experienced, long-term immunity that can be found at low frequencies in circulating peripheral blood mononuclear cells (PBMCs). Assessing the genotype, clonal frequency, quality, and function of antibodies resulting from an individual's persistent memory B cell repertoire can help inform the success or failure of immune protection. Using in vitro polyclonal stimulation, we functionally expand the memory repertoire from PBMCs and clonally map monoclonal antibodies from this population. We show that combining deep sequencing of stimulated memory B cell repertoires with retrieving single antigen-specific cells is a promising approach in evaluating the latent, functional B cell memory in PBMCs.
Keywords: PBMC; antibody; clonal families; memory B cell; repertoire; sequencing.
Figures


Similar articles
-
Insights From Analysis of Human Antigen-Specific Memory B Cell Repertoires.Front Immunol. 2019 Jan 15;9:3064. doi: 10.3389/fimmu.2018.03064. eCollection 2018. Front Immunol. 2019. PMID: 30697210 Free PMC article. Review.
-
Distinct kinetics of memory B-cell and plasma-cell responses in peripheral blood following a blood-stage Plasmodium chabaudi infection in mice.PLoS One. 2010 Nov 23;5(11):e15007. doi: 10.1371/journal.pone.0015007. PLoS One. 2010. PMID: 21124900 Free PMC article.
-
Expansion of IgG+ B-cells during mitogen stimulation for memory B-cell ELISpot analysis is influenced by size and composition of the B-cell pool.PLoS One. 2014 Jul 22;9(7):e102885. doi: 10.1371/journal.pone.0102885. eCollection 2014. PLoS One. 2014. PMID: 25050555 Free PMC article.
-
Protective long-term antibody memory by antigen-driven and T help-dependent differentiation of long-lived memory B cells to short-lived plasma cells independent of secondary lymphoid organs.Proc Natl Acad Sci U S A. 2000 Nov 21;97(24):13263-8. doi: 10.1073/pnas.230417497. Proc Natl Acad Sci U S A. 2000. PMID: 11069289 Free PMC article.
-
Non-classical B Cell Memory of Allergic IgE Responses.Front Immunol. 2019 Apr 26;10:715. doi: 10.3389/fimmu.2019.00715. eCollection 2019. Front Immunol. 2019. PMID: 31105687 Free PMC article. Review.
Cited by
-
Combining phage display with SMRTbell next-generation sequencing for the rapid discovery of functional scFv fragments.MAbs. 2021 Jan-Dec;13(1):1864084. doi: 10.1080/19420862.2020.1864084. MAbs. 2021. PMID: 33382949 Free PMC article.
-
Flow cytometric protocol to characterize human memory B cells directed against SARS-CoV-2 spike protein antigens.STAR Protoc. 2022 Dec 16;3(4):101902. doi: 10.1016/j.xpro.2022.101902. Epub 2022 Nov 15. STAR Protoc. 2022. PMID: 36595922 Free PMC article.
-
Navigating the Landscape of B Cell Mediated Immunity and Antibody Monitoring in SARS-CoV-2 Vaccine Efficacy: Tools, Strategies and Clinical Trial Insights.Vaccines (Basel). 2024 Sep 24;12(10):1089. doi: 10.3390/vaccines12101089. Vaccines (Basel). 2024. PMID: 39460256 Free PMC article. Review.
-
Advances in antibody discovery from human BCR repertoires.Front Bioinform. 2022 Oct 20;2:1044975. doi: 10.3389/fbinf.2022.1044975. eCollection 2022. Front Bioinform. 2022. PMID: 36338807 Free PMC article. Review.
-
Targeting Neoepitopes to Treat Solid Malignancies: Immunosurgery.Front Immunol. 2021 Jul 15;12:592031. doi: 10.3389/fimmu.2021.592031. eCollection 2021. Front Immunol. 2021. PMID: 34335558 Free PMC article. Review.
References
-
- Abbas AK, Lichtman AH, Pillai S. Cellular and Molecular Immunology, 9th ed. Philadelphia, PA: Elsevier Health Sciences; (2017). ISBN: 9780323523233.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

