SAMHD1 Modulates Early Steps during Human Cytomegalovirus Infection by Limiting NF-κB Activation
- PMID: 31291579
- PMCID: PMC6662646
- DOI: 10.1016/j.celrep.2019.06.027
SAMHD1 Modulates Early Steps during Human Cytomegalovirus Infection by Limiting NF-κB Activation
Abstract
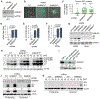
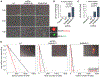
Cellular SAMHD1 inhibits replication of many viruses by limiting intracellular deoxynucleoside triphosphate (dNTP) pools. We investigate the influence of SAMHD1 on human cytomegalovirus (HCMV). During HCMV infection, we observe SAMHD1 induction, accompanied by phosphorylation via viral kinase UL97. SAMHD1 depletion increases HCMV replication in permissive fibroblasts and conditionally permissive myeloid cells. We show this is due to enhanced gene expression from the major immediate-early (MIE) promoter and is independent of dNTP levels. SAMHD1 suppresses innate immune responses by inhibiting nuclear factor κB (NF-κB) activation. We show that SAMHD1 regulates the HCMV MIE promoter through NF-κB activation. Chromatin immunoprecipitation reveals increased RELA and RNA polymerase II on the HCMV MIE promoter in the absence of SAMHD1. Our studies reveal a mechanism of HCMV virus restriction by SAMHD1 and show how SAMHD1 deficiency activates an innate immune pathway that paradoxically results in increased viral replication through transcriptional activation of the HCMV MIE gene promoter.
Keywords: HCMV; NF-κB; SAMHD1; human cytomegalovirus; virus restriction.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


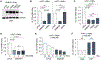

Similar articles
-
Human cytomegalovirus overcomes SAMHD1 restriction in macrophages via pUL97.Nat Microbiol. 2019 Dec;4(12):2260-2272. doi: 10.1038/s41564-019-0557-8. Epub 2019 Sep 23. Nat Microbiol. 2019. PMID: 31548682
-
TRAF6 and TAK1 Contribute to SAMHD1-Mediated Negative Regulation of NF-κB Signaling.J Virol. 2021 Jan 13;95(3):e01970-20. doi: 10.1128/JVI.01970-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177202 Free PMC article.
-
SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-κB and interferon pathways.Proc Natl Acad Sci U S A. 2018 Apr 17;115(16):E3798-E3807. doi: 10.1073/pnas.1801213115. Epub 2018 Apr 2. Proc Natl Acad Sci U S A. 2018. PMID: 29610295 Free PMC article.
-
SAMHD1 Functions and Human Diseases.Viruses. 2020 Mar 31;12(4):382. doi: 10.3390/v12040382. Viruses. 2020. PMID: 32244340 Free PMC article. Review.
-
Intrinsic Immune Mechanisms Restricting Human Cytomegalovirus Replication.Viruses. 2021 Jan 26;13(2):179. doi: 10.3390/v13020179. Viruses. 2021. PMID: 33530304 Free PMC article. Review.
Cited by
-
SAMHD1 phosphorylation and cytoplasmic relocalization after human cytomegalovirus infection limits its antiviral activity.PLoS Pathog. 2020 Sep 28;16(9):e1008855. doi: 10.1371/journal.ppat.1008855. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32986788 Free PMC article.
-
Reprogramming of cellular metabolic pathways by human oncogenic viruses.Curr Opin Virol. 2019 Dec;39:60-69. doi: 10.1016/j.coviro.2019.11.002. Epub 2019 Nov 22. Curr Opin Virol. 2019. PMID: 31766001 Free PMC article. Review.
-
Schlafens Can Put Viruses to Sleep.Viruses. 2022 Feb 21;14(2):442. doi: 10.3390/v14020442. Viruses. 2022. PMID: 35216035 Free PMC article. Review.
-
Human cytomegalovirus induces neuronal gene expression for viral maturation.bioRxiv [Preprint]. 2024 Jun 13:2024.06.13.598910. doi: 10.1101/2024.06.13.598910. bioRxiv. 2024. PMID: 38915666 Free PMC article. Preprint.
-
Targeting SAMHD1: To overcome multiple anti-cancer drugs resistance in hematological malignancies.Genes Dis. 2022 Jun 26;10(3):891-900. doi: 10.1016/j.gendis.2022.06.001. eCollection 2023 May. Genes Dis. 2022. PMID: 37396510 Free PMC article. Review.
References
-
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, and Hornung V (2014). TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol 192, 5993–5997. - PubMed
-
- Adler M, Tavalai N, Müller R, and Stamminger T (2011). Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol 92, 1532–1538. - PubMed
-
- Ahn JH, and Hayward GS (2000). Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274, 39–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

